+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9693 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an alphavirus, Sindbis virus | |||||||||
 Map data Map data | Cryo-EM structure of an alphavirus, Sindbis virus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Alphavirus / Sindbis virus / Glycoprotein / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral capsid / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis ...viral capsid / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Sindbis virus Sindbis virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Zhang X / Ma J | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Implication for alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure. Authors: Lihong Chen / Ming Wang / Dongjie Zhu / Zhenzhao Sun / Jun Ma / Jinglin Wang / Lingfei Kong / Shida Wang / Zaisi Liu / Lili Wei / Yuwen He / Jingfei Wang / Xinzheng Zhang /  Abstract: Alphaviruses are enveloped RNA viruses that contain several human pathogens. Due to intrinsic heterogeneity of alphavirus particles, a high resolution structure of the virion is currently lacking. ...Alphaviruses are enveloped RNA viruses that contain several human pathogens. Due to intrinsic heterogeneity of alphavirus particles, a high resolution structure of the virion is currently lacking. Here we provide a 3.5 Å cryo-EM structure of Sindbis virus, using block based reconstruction method that overcomes the heterogeneity problem. Our structural analysis identifies a number of conserved residues that play pivotal roles in the virus life cycle. We identify a hydrophobic pocket in the subdomain D of E2 protein that is stabilized by an unknown pocket factor near the viral membrane. Residues in the pocket are conserved in different alphaviruses. The pocket strengthens the interactions of the E1/E2 heterodimer and may facilitate virus assembly. Our study provides structural insights into alphaviruses that may inform the design of drugs and vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9693.map.gz emd_9693.map.gz | 360.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9693-v30.xml emd-9693-v30.xml emd-9693.xml emd-9693.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9693.png emd_9693.png | 297.2 KB | ||
| Filedesc metadata |  emd-9693.cif.gz emd-9693.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9693 http://ftp.pdbj.org/pub/emdb/structures/EMD-9693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9693 | HTTPS FTP |
-Validation report
| Summary document |  emd_9693_validation.pdf.gz emd_9693_validation.pdf.gz | 723.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9693_full_validation.pdf.gz emd_9693_full_validation.pdf.gz | 722.7 KB | Display | |
| Data in XML |  emd_9693_validation.xml.gz emd_9693_validation.xml.gz | 8.6 KB | Display | |
| Data in CIF |  emd_9693_validation.cif.gz emd_9693_validation.cif.gz | 10 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9693 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9693 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9693 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9693 | HTTPS FTP |
-Related structure data
| Related structure data |  6immMC  9692C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9693.map.gz / Format: CCP4 / Size: 744.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9693.map.gz / Format: CCP4 / Size: 744.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of an alphavirus, Sindbis virus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
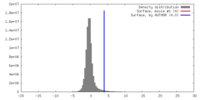
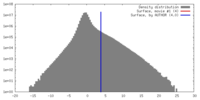
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sindbis virus
| Entire | Name:  Sindbis virus Sindbis virus |
|---|---|
| Components |
|
-Supramolecule #1: Sindbis virus
| Supramolecule | Name: Sindbis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 11034 / Sci species name: Sindbis virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sindbis virus Sindbis virus |
| Molecular weight | Theoretical: 47.186434 KDa |
| Sequence | String: FEHATTVPNV PRIPYKALVE RAGYAPLNLE ITVMSSELIP STNLEYVTCK YTTVVPSPKV KCCGTLECSS ARHADYNCKV FGGVYPFMW GGAQCFCDSE NSQMSEAYVE FSADCAADHA QAVKVHTAAL KAGLRIVYGN TTSMLDVYVN GVTPGTSKDL K VIAGPISA ...String: FEHATTVPNV PRIPYKALVE RAGYAPLNLE ITVMSSELIP STNLEYVTCK YTTVVPSPKV KCCGTLECSS ARHADYNCKV FGGVYPFMW GGAQCFCDSE NSQMSEAYVE FSADCAADHA QAVKVHTAAL KAGLRIVYGN TTSMLDVYVN GVTPGTSKDL K VIAGPISA AYTPFDHKVI IHKGKVYNYD FPEYGAMKPG AFGDIQATSL TSNDLIANTD IRLLKPSAKN VHVPYTQAAS GF EMWKNNS GRPLQETAPF GCQIAVNPLR AVDCAYGNIP ISLDIPNAAF VRVSDAPLVT ALKCEVGECV YSADFGGIAT LQY SSDREG QCSVHSHSST ATLQESTVHV LQKGGATIHF STASPQANFI VSLCGKKTTC NAECKPPADH IVNVPHKNDQ EFQA AVSQT SWSWLFALFG GASSLLVIGV MIFACSALLT STRR |
-Macromolecule #2: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sindbis virus Sindbis virus |
| Molecular weight | Theoretical: 43.184387 KDa |
| Sequence | String: FTLTSPYLGT CSYCHHTEPC FSPVKIEQVW DEADDNTIRI QTSAQFGYDQ SGAASVNKYR IMSLKQDHTI EEGSMDAIKI STSGPCRRL NHKGYFLLAK CPPGDSVTVS ISAGDSATSC TLARKVKPKF VGREKYDLPP VHGKKIPCYI YDRLKETSAG Y ITMHRPGP ...String: FTLTSPYLGT CSYCHHTEPC FSPVKIEQVW DEADDNTIRI QTSAQFGYDQ SGAASVNKYR IMSLKQDHTI EEGSMDAIKI STSGPCRRL NHKGYFLLAK CPPGDSVTVS ISAGDSATSC TLARKVKPKF VGREKYDLPP VHGKKIPCYI YDRLKETSAG Y ITMHRPGP HAYATYLEES SGKVYAKPPS GKNITYKSDQ TKWVFNSPDL IRHADHTAQG KMHLPFKLVP STCLVPLAHV PQ VVHGFKH ISLQLDTDHL TLLTTRRLGE KPEPTSEWII GKTVRNFSVG RDGFEYIWGN HEPVRVWAQE SAPGDPHGWP HEI VQHYYH RHPVYTVMIL VAATLAIVLG VSVASVCVCR ARRECLTPYA LAPNAVVPTS IALLCCIRPT SA |
-Macromolecule #3: Assembly protein E3
| Macromolecule | Name: Assembly protein E3 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sindbis virus Sindbis virus |
| Molecular weight | Theoretical: 7.480542 KDa |
| Sequence | String: SAAPLVAAMC ILGNMTFPCN QPPTCYSREP ARALDILEAN VDSAAYDDLM RAVLRCTPSS RAKRNITDD |
-Macromolecule #4: Octadecane
| Macromolecule | Name: Octadecane / type: ligand / ID: 4 / Number of copies: 1 / Formula: 8K6 |
|---|---|
| Molecular weight | Theoretical: 254.494 Da |
| Chemical component information |  ChemComp-8K6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 29974 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)