+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9654 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of type III-A Csm-NTR complex | |||||||||
 Map data Map data | Cryo-EM structure of type III-A Csm-NTR complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Csm complex / Type III-A / CRISPR-Cas system / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationexonuclease activity / transferase activity / endonuclease activity / defense response to virus / RNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Streptococcus thermophilus ND03 (bacteria) / Streptococcus thermophilus ND03 (bacteria) /  Streptococcus thermophilus (bacteria) Streptococcus thermophilus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.16 Å | |||||||||
 Authors Authors | You L / Ma J | |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference. Authors: Lilan You / Jun Ma / Jiuyu Wang / Daria Artamonova / Min Wang / Liang Liu / Hua Xiang / Konstantin Severinov / Xinzheng Zhang / Yanli Wang /   Abstract: Csm, a type III-A CRISPR-Cas interference complex, is a CRISPR RNA (crRNA)-guided RNase that also possesses target RNA-dependent DNase and cyclic oligoadenylate (cOA) synthetase activities. However, ...Csm, a type III-A CRISPR-Cas interference complex, is a CRISPR RNA (crRNA)-guided RNase that also possesses target RNA-dependent DNase and cyclic oligoadenylate (cOA) synthetase activities. However, the structural features allowing target RNA-binding-dependent activation of DNA cleavage and cOA generation remain unknown. Here, we report the structure of Csm in complex with crRNA together with structures of cognate or non-cognate target RNA bound Csm complexes. We show that depending on complementarity with the 5' tag of crRNA, the 3' anti-tag region of target RNA binds at two distinct sites of the Csm complex. Importantly, the interaction between the non-complementary anti-tag region of cognate target RNA and Csm1 induces a conformational change at the Csm1 subunit that allosterically activates DNA cleavage and cOA generation. Together, our structural studies provide crucial insights into the mechanistic processes required for crRNA-meditated sequence-specific RNA cleavage, RNA target-dependent non-specific DNA cleavage, and cOA generation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9654.map.gz emd_9654.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9654-v30.xml emd-9654-v30.xml emd-9654.xml emd-9654.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9654.png emd_9654.png | 64.1 KB | ||
| Filedesc metadata |  emd-9654.cif.gz emd-9654.cif.gz | 6.8 KB | ||
| Others |  emd_9654_additional.map.gz emd_9654_additional.map.gz | 28.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9654 http://ftp.pdbj.org/pub/emdb/structures/EMD-9654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9654 | HTTPS FTP |
-Related structure data
| Related structure data |  6iflMC  9653C  9655C  9656C  9657C  9658C  9659C  9660C  6ifkC  6ifnC  6ifrC  6ifuC  6ifyC  6ifzC  6ig0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9654.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9654.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of type III-A Csm-NTR complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
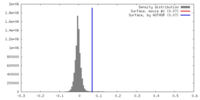
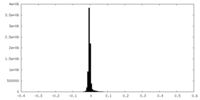
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Cryo-EM structure of type III-A Csm-NTR complex,subunit Csm1
| File | emd_9654_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of type III-A Csm-NTR complex,subunit Csm1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
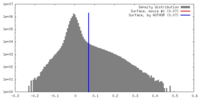
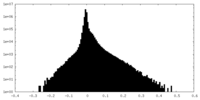
| Density Histograms |
- Sample components
Sample components
-Entire : Csm-CTR1 complex, AMPPNP bound
| Entire | Name: Csm-CTR1 complex, AMPPNP bound |
|---|---|
| Components |
|
-Supramolecule #1: Csm-CTR1 complex, AMPPNP bound
| Supramolecule | Name: Csm-CTR1 complex, AMPPNP bound / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: type III-A CRISPR-Cas interference complex |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 87 KDa |
-Supramolecule #2: Csm1
| Supramolecule | Name: Csm1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: type III-A CRISPR-Cas interference complex subunit Csm1 |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
-Macromolecule #1: Type III-A CRISPR-associated protein Csm1
| Macromolecule | Name: Type III-A CRISPR-associated protein Csm1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 86.976734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKEKIDLFY GALLHNIGKV IQRATGERKK HALVGADWFD EIADNQVISD QIRYHMANYQ SDKLGNDHLA YITYIADNIA SGVDRRQSN EESDEDTSAK IWDTYTNQAD IFNVFGAQTD KRYFKPTVLN LKSKPNFASA TYEPFSKGDY AAIATRIKNE L AEFEFNQV ...String: MKKEKIDLFY GALLHNIGKV IQRATGERKK HALVGADWFD EIADNQVISD QIRYHMANYQ SDKLGNDHLA YITYIADNIA SGVDRRQSN EESDEDTSAK IWDTYTNQAD IFNVFGAQTD KRYFKPTVLN LKSKPNFASA TYEPFSKGDY AAIATRIKNE L AEFEFNQV QIDSLLNLFE ATLSFVPSST NTKEIADISL ADHSRLTAAF ALAIYDYLED KGRHNYKEDL FTKVSAFYEE EA FLLASFD LSGIQDFIYN INIATNGAAK QLKARSLYLD FMSEYIADSL LDKLGLNRAN MLYVGGGHAY FVLANTEKTV ETL VQFEKD FNQFLLANFQ TRLYVAFGWG SFAAKDIMSE LNSPESYRQV YQKASRMISK KKISRYDYQT LMLLNRGGKS SERE CEICH SVENLVSYHD QKVCDICRGL YQFSKEIAHD HFIITENEGL PIGPNACLKG VAFEKLSQEA FSRVYVKNDY KAGTV KATH VFVGDYQCDE IYNYAALSKN ENGLGIKRLA VVRLDVDDLG AAFMAGFSQQ GNGQYSTLSR SATFSRSMSL FFKVYI NQF ASDKKLSIIY AGGDDVFAIG SWQDIIAFTV ELRENFIKWT NGKLTLSAGI GLFADKTPIS LMAHQTGELE EAAKGNE KD SISLFSSDYT FKFDRFITNV YDDKLEQIRY FFNHQDERGK NFIYKLIELL RNHDRMNMAR LAYYLTRLEE LTRETDRD K FKTFKNLFYS WYTNKNDKDR KEAELALLLY IYEIRKD UniProtKB: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A) |
-Macromolecule #2: Type III-A CRISPR-associated protein Csm2
| Macromolecule | Name: Type III-A CRISPR-associated protein Csm2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 14.848145 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTILTDENYV DIAEKAILKL ERNTRNRKNP DAFFLTTSKL RNLLSLTSTL FDESKVKEYD ALLDRIAYLR VQFVYQAGRE IAVKDLIEK AQILEALKEI KDRETLQRFC RYMEALVAYF KFYGGKD UniProtKB: CRISPR system Cms protein Csm2 |
-Macromolecule #3: Type III-A CRISPR-associated RAMP protein Csm3
| Macromolecule | Name: Type III-A CRISPR-associated RAMP protein Csm3 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 24.584855 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTFAKIKFSA QIRLETGLHI GGSDAFAAIG AINSPVIKDP ITNLPIIPGS SLKGKMRTLL AKVYNEKVAE KPSDDSDILS RLFGNSKDK RFKMGRLIFR DAFLSNADEL DSLGVRSYTE VKFENTIDRI TAEANPRQIE RAIRNSTFDF ELIYEITDEN E NQVEEDFK ...String: MTFAKIKFSA QIRLETGLHI GGSDAFAAIG AINSPVIKDP ITNLPIIPGS SLKGKMRTLL AKVYNEKVAE KPSDDSDILS RLFGNSKDK RFKMGRLIFR DAFLSNADEL DSLGVRSYTE VKFENTIDRI TAEANPRQIE RAIRNSTFDF ELIYEITDEN E NQVEEDFK VIRDGLKLLE LDYLGGSGSR GYGKVAFENL KATTVFGNYD VKTLNELLTA EV UniProtKB: CRISPR system Cms endoribonuclease Csm3 |
-Macromolecule #4: Type III-A CRISPR-associated RAMP protein Csm4
| Macromolecule | Name: Type III-A CRISPR-associated RAMP protein Csm4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 33.828984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTYKLYIMTF QNAHFGSGTL DSSKLTFSAD RIFSALVLEA LKMGKLDAFL AEANQDKFTL TDAFPFQFGP FLPKPIGYPK HDQIDQSVD VKEVRRQAKL SKKLQFLALE NVDDYLNGEL FENEEHAVID TVTKNQPHKD DNLYQVATTR FSNDTSLYVI A NESDLLNE ...String: MTYKLYIMTF QNAHFGSGTL DSSKLTFSAD RIFSALVLEA LKMGKLDAFL AEANQDKFTL TDAFPFQFGP FLPKPIGYPK HDQIDQSVD VKEVRRQAKL SKKLQFLALE NVDDYLNGEL FENEEHAVID TVTKNQPHKD DNLYQVATTR FSNDTSLYVI A NESDLLNE LMSSLQYSGL GGKRSSGFGR FELDIQNIPL ELSDRLTKNH SDKVMSLTTA LPVDADLEEA MEDGHYLLTK SS GFAFSHA TNENYRKQDL YKFASGSTFS KTFEGQIVDV RPLDFPHAVL NYAKPLFFKL EV UniProtKB: CRISPR system Cms protein Csm4 |
-Macromolecule #5: Type III-A CRISPR-associated RAMP protein Csm5
| Macromolecule | Name: Type III-A CRISPR-associated RAMP protein Csm5 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus (bacteria) Streptococcus thermophilus (bacteria) |
| Molecular weight | Theoretical: 41.102113 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKNDYRTFKL SLLTLAPIHI GNGEKYTSRE FIYENKKFYF PDMGKFYNKM VEKRLAEKFE AFLIQTRPNA RNNRLISFLN DNRIAERSF GGYSISETGL ESDKNPNSAG AINEVNKFIR DAFGNPYIPG SSLKGAIRTI LMNTTPKWNN ENAVNDFGRF P KENKNLIP ...String: MKNDYRTFKL SLLTLAPIHI GNGEKYTSRE FIYENKKFYF PDMGKFYNKM VEKRLAEKFE AFLIQTRPNA RNNRLISFLN DNRIAERSF GGYSISETGL ESDKNPNSAG AINEVNKFIR DAFGNPYIPG SSLKGAIRTI LMNTTPKWNN ENAVNDFGRF P KENKNLIP WGPKKGKEYD DLFNAIRVSD SKPFDNKSLI LVQKWDYSAK TNKAKPLPLY RESISPLTKI EFEITTTTDE AG RLIEELG KRAQAFYKDY KAFFLSEFPD DKIQANLQYP IYLGAGSGAW TKTLFKQADG ILQRRYSRMK TKMVKKGVLK LTK APLKTV KIPSGNHSLV KNHESFYEMG KANFMIKEID K UniProtKB: CRISPR system Cms protein Csm5 |
-Macromolecule #6: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 11.39277 KDa |
| Sequence | String: ACGGAAACGC UUUCUAGCUC GCUAUAAUUA CCCAUU |
-Macromolecule #7: NTR
| Macromolecule | Name: NTR / type: rna / ID: 7 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus ND03 (bacteria) Streptococcus thermophilus ND03 (bacteria) |
| Molecular weight | Theoretical: 13.937306 KDa |
| Sequence | String: GGUAGGAAUG GGUAAUUAUA GCGAGCUAGA AAGCGUUUCC GUC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 60 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.16 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 133127 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)