+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9327 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacillus PS3 ATP synthase membrane region | |||||||||
 Map data Map data | ATP synthase membrane region | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / lipid binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
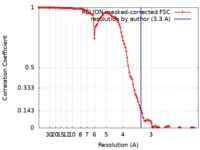
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Guo H / Rubinstein JL | |||||||||
| Funding support |  Canada, Canada,  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Structure of a bacterial ATP synthase. Authors: Hui Guo / Toshiharu Suzuki / John L Rubinstein /   Abstract: ATP synthases produce ATP from ADP and inorganic phosphate with energy from a transmembrane proton motive force. Bacterial ATP synthases have been studied extensively because they are the simplest ...ATP synthases produce ATP from ADP and inorganic phosphate with energy from a transmembrane proton motive force. Bacterial ATP synthases have been studied extensively because they are the simplest form of the enzyme and because of the relative ease of genetic manipulation of these complexes. We expressed the PS3 ATP synthase in , purified it, and imaged it by cryo-EM, allowing us to build atomic models of the complex in three rotational states. The position of subunit shows how it is able to inhibit ATP hydrolysis while allowing ATP synthesis. The architecture of the membrane region shows how the simple bacterial ATP synthase is able to perform the same core functions as the equivalent, but more complicated, mitochondrial complex. The structures reveal the path of transmembrane proton translocation and provide a model for understanding decades of biochemical analysis interrogating the roles of specific residues in the enzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9327.map.gz emd_9327.map.gz | 116.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9327-v30.xml emd-9327-v30.xml emd-9327.xml emd-9327.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9327_fsc.xml emd_9327_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_9327.png emd_9327.png | 107.7 KB | ||
| Masks |  emd_9327_msk_1.map emd_9327_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9327.cif.gz emd-9327.cif.gz | 6.2 KB | ||
| Others |  emd_9327_half_map_1.map.gz emd_9327_half_map_1.map.gz emd_9327_half_map_2.map.gz emd_9327_half_map_2.map.gz | 97.8 MB 97.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9327 http://ftp.pdbj.org/pub/emdb/structures/EMD-9327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9327 | HTTPS FTP |
-Related structure data
| Related structure data |  6n2dMC  9333C  9334C  9335C  9336C  9337C  9338C  6n2yC  6n2zC  6n30C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9327.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9327.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATP synthase membrane region | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
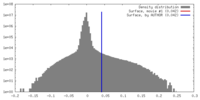
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9327_msk_1.map emd_9327_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
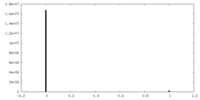
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: ATP synthase membrane region
| File | emd_9327_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATP synthase membrane region | ||||||||||||
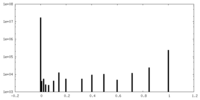
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: ATP synthase membrane region
| File | emd_9327_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATP synthase membrane region | ||||||||||||
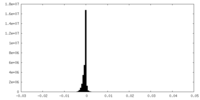
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Membrane-embedded region of Bacillus PS3 ATP synthase
| Entire | Name: Membrane-embedded region of Bacillus PS3 ATP synthase |
|---|---|
| Components |
|
-Supramolecule #1: Membrane-embedded region of Bacillus PS3 ATP synthase
| Supramolecule | Name: Membrane-embedded region of Bacillus PS3 ATP synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 110 kDa/nm |
-Macromolecule #1: ATP synthase subunit b
| Macromolecule | Name: ATP synthase subunit b / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.249148 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EAAHGISGGT IIYQLLMFII LLALLRKFAW QPLMNIMKQR EEHIANEIDQ AEKRRQEAEK LLEEQRELMK QSRQEAQALI ENARKLAEE QKEQIVASAR AEAERVKETA KKEIEREKEQ AMAALREQVA SLSVLIASKV IEKELTEQDQ RKLIEAYIKD V QEVGGAR |
-Macromolecule #2: ATP synthase subunit a
| Macromolecule | Name: ATP synthase subunit a / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.44932 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEHKAPLVEF LGLTFNLSDM LMITITCLIV FIIAVAATRS LQLRPTGMQN FMEWVFDFVR GIINSTMDWQ TGGRFLTLGV TLIMYVFVA NMLGLPFSVH VNGELWWKSP TADATVTLTL AVMVVALTHY YGVKMKGASD YLRDYTRPVA WLFPLKIIEE F ANTLTLGL ...String: MEHKAPLVEF LGLTFNLSDM LMITITCLIV FIIAVAATRS LQLRPTGMQN FMEWVFDFVR GIINSTMDWQ TGGRFLTLGV TLIMYVFVA NMLGLPFSVH VNGELWWKSP TADATVTLTL AVMVVALTHY YGVKMKGASD YLRDYTRPVA WLFPLKIIEE F ANTLTLGL RLFGNIYAGE ILLGLLASLG THYGVLGAVG AAIPMMVWQA FSIFVGTIQA FIFTMLTMVY MAHKVSHDH |
-Macromolecule #3: ATP synthase subunit c
| Macromolecule | Name: ATP synthase subunit c / type: protein_or_peptide / ID: 3 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.33778 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLGVLAAAI AVGLGALGAG IGNGLIVSRT IEGIARQPEL RPVLQTTMFI GVALVEALPI IGVVFSFIYL GR UniProtKB: ATP synthase subunit c |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Material: COPPER/RHODIUM / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average exposure time: 60.0 sec. / Average electron dose: 0.71 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 132075 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 91.12 |
|---|---|
| Output model |  PDB-6n2d: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)