[English] 日本語
 Yorodumi
Yorodumi- EMDB-8397: In situ structures of the genome and genome-delivery apparatus in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8397 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structures of the genome and genome-delivery apparatus in ssRNA bacteriophage MS2 | |||||||||
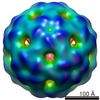
 Map data Map data | Asymmetric cryoEM reconstruction of Enterobacteria Phage MS2 at 3.6 Angstroms resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | asymmetric cryoEM reconstruction / ssRNA genome structure / genome-delivery apparatus / genome-capsid interactions / viral protein-rna complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral genome circularization / virion attachment to host cell pilus / negative regulation of viral translation / T=3 icosahedral viral capsid / virion component / regulation of translation / structural molecule activity / RNA binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Dai XH / Li ZH | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Authors: Xinghong Dai / Zhihai Li / Mason Lai / Sara Shu / Yushen Du / Z Hong Zhou / Ren Sun /   Abstract: Packaging of the genome into a protein capsid and its subsequent delivery into a host cell are two fundamental processes in the life cycle of a virus. Unlike double-stranded DNA viruses, which pump ...Packaging of the genome into a protein capsid and its subsequent delivery into a host cell are two fundamental processes in the life cycle of a virus. Unlike double-stranded DNA viruses, which pump their genome into a preformed capsid, single-stranded RNA (ssRNA) viruses, such as bacteriophage MS2, co-assemble their capsid with the genome; however, the structural basis of this co-assembly is poorly understood. MS2 infects Escherichia coli via the host 'sex pilus' (F-pilus); it was the first fully sequenced organism and is a model system for studies of translational gene regulation, RNA-protein interactions, and RNA virus assembly. Its positive-sense ssRNA genome of 3,569 bases is enclosed in a capsid with one maturation protein monomer and 89 coat protein dimers arranged in a T = 3 icosahedral lattice. The maturation protein is responsible for attaching the virus to an F-pilus and delivering the viral genome into the host during infection, but how the genome is organized and delivered is not known. Here we describe the MS2 structure at 3.6 Å resolution, determined by electron-counting cryo-electron microscopy (cryoEM) and asymmetric reconstruction. We traced approximately 80% of the backbone of the viral genome, built atomic models for 16 RNA stem-loops, and identified three conserved motifs of RNA-coat protein interactions among 15 of these stem-loops with diverse sequences. The stem-loop at the 3' end of the genome interacts extensively with the maturation protein, which, with just a six-helix bundle and a six-stranded β-sheet, forms a genome-delivery apparatus and joins 89 coat protein dimers to form a capsid. This atomic description of genome-capsid interactions in a spherical ssRNA virus provides insight into genome delivery via the host sex pilus and mechanisms underlying ssRNA-capsid co-assembly, and inspires speculation about the links between nucleoprotein complexes and the origins of viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8397.map.gz emd_8397.map.gz | 109 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8397-v30.xml emd-8397-v30.xml emd-8397.xml emd-8397.xml | 27.7 KB 27.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8397.png emd_8397.png | 281.1 KB | ||
| Filedesc metadata |  emd-8397.cif.gz emd-8397.cif.gz | 8.5 KB | ||
| Others |  emd_8397_additional.map.gz emd_8397_additional.map.gz | 58 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8397 http://ftp.pdbj.org/pub/emdb/structures/EMD-8397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8397 | HTTPS FTP |
-Validation report
| Summary document |  emd_8397_validation.pdf.gz emd_8397_validation.pdf.gz | 679.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8397_full_validation.pdf.gz emd_8397_full_validation.pdf.gz | 679.2 KB | Display | |
| Data in XML |  emd_8397_validation.xml.gz emd_8397_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_8397_validation.cif.gz emd_8397_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8397 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8397 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8397 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8397 | HTTPS FTP |
-Related structure data
| Related structure data |  5tc1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8397.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8397.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric cryoEM reconstruction of Enterobacteria Phage MS2 at 3.6 Angstroms resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
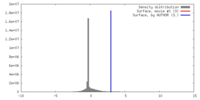
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Asymmetric cryoEM reconstruction of Enterobacteria Phage MS2 low-pass...
| File | emd_8397_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric cryoEM reconstruction of Enterobacteria Phage MS2 low-pass filtered to 6 Angstroms resolution to visualize the ssRNA genome. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage MS2
| Entire | Name:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage MS2
| Supramolecule | Name: Enterobacteria phage MS2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: The viral stock was obtained from ATCC (ATCC number 15597-B1) and cultured in Escherichia coli strain C-3000 (ATCC 15597). NCBI-ID: 329852 / Sci species name: Enterobacteria phage MS2 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Theoretical: 3.6 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 270.0 Å / T number (triangulation number): 3 |
-Supramolecule #2: capsid shell of Enterobacteria Phage MS2
| Supramolecule | Name: capsid shell of Enterobacteria Phage MS2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 Details: The capsid of MS2 is composed of 178 copies of the coat protein and one single copy of the maturation protein. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 2.5 MDa |
-Supramolecule #3: coat protein of Enterobacteria Phage MS2
| Supramolecule | Name: coat protein of Enterobacteria Phage MS2 / type: complex / ID: 3 / Parent: 2 / Macromolecule list: #1 Details: The 178 copies of the coat protein are organized as 89 dimers in a T=3 icosahedral lattice. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 14 KDa |
-Supramolecule #4: maturation protein of Enterobacteria Phage MS2
| Supramolecule | Name: maturation protein of Enterobacteria Phage MS2 / type: complex / ID: 4 / Parent: 2 / Macromolecule list: #2 Details: The single copy of maturation protein in the capsid shell of MS2 is located at one of the 2-fold symmetry axes and it replaces a coat protein dimer at this position. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 44 KDa |
-Supramolecule #5: the ssRNA genome of Enterobacteria Phage MS2
| Supramolecule | Name: the ssRNA genome of Enterobacteria Phage MS2 / type: complex / ID: 5 / Parent: 1 / Macromolecule list: #3 Details: The genome of MS2 is a single-stranded RNA with 3569 bases. Our asymmetric cryoEM reconstruction of the MS2 virion shows that its ssRNA genome is well organized and has multiple contacts ...Details: The genome of MS2 is a single-stranded RNA with 3569 bases. Our asymmetric cryoEM reconstruction of the MS2 virion shows that its ssRNA genome is well organized and has multiple contacts with the capsid shell via tens of RNA stem-loop structures. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 1.1 MDa |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 Details: The capsid of MS2 contains 178 copies of the coat protein arranged as 89 dimers in a T=3 icosahedral lattice. Structure of the capsid has been solved by crystallography with icosahedral ...Details: The capsid of MS2 contains 178 copies of the coat protein arranged as 89 dimers in a T=3 icosahedral lattice. Structure of the capsid has been solved by crystallography with icosahedral symmetry applied (PDB ID 2MS2, also included as chains A, B, C in this model). Chains D, E, F, G, H are coat proteins that have slightly different structures in the asymmetric cryoEM reconstruction compared to the crystallographic structure. Structures of the other 170 copies of the coat protein are the same with 2MS2, and thus are not included due to the limited number of chain IDs. Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 13.869659 KDa |
| Sequence | String: MASNFTQFVL VDNGGTGDVT VAPSNFANGV AEWISSNSRS QAYKVTCSVR QSSAQNRKYT IKVEVPKVAT QTVGGVELPV AAWRSYLNM ELTIPIFATN SDCELIVKAM QGLLKDGNPI PSAIAANSGI Y UniProtKB: Capsid protein |
-Macromolecule #2: Maturation protein
| Macromolecule | Name: Maturation protein / type: protein_or_peptide / ID: 2 Details: The capsid of MS2 contains a single copy of the maturation protein. Our structure shows that it replaces a coat protein dimer at one of the 2-fold icosahedral symmetry axes. Function of the ...Details: The capsid of MS2 contains a single copy of the maturation protein. Our structure shows that it replaces a coat protein dimer at one of the 2-fold icosahedral symmetry axes. Function of the maturation protein is to attach the MS2 virion to the host (E. coli) F-pili and deliver the ssRNA viral genome into the host during infection. of the icosahedral capsid. 178 copies of the coat protein arranged as 89 dimers in a T=3 icosahedral lattice. Structure of the capsid has been solved by crystallography with icosahedral symmetry applied (PDB ID 2MS2, also included as chains A, B, C in this model). Chains D, E, F, G, H are coat proteins that have slightly different structures in the asymmetric cryoEM reconstruction compared to the crystallographic structure. Structures of the other 170 copies of the coat protein are the same with 2MS2, and thus are not included due to the limited number of chain IDs. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 44.030934 KDa |
| Sequence | String: MRAFSTLDRE NETFVPSVRV YADGETEDNS FSLKYRSNWT PGRFNSTGAK TKQWHYPSPY SRGALSVTSI DQGAYKRSGS SWGRPYEEK AGFGFSLDAR SCYSLFPVSQ NLTYIEVPQN VANRASTEVL QKVTQGNFNL GVALAEARST ASQLATQTIA L VKAYTAAR ...String: MRAFSTLDRE NETFVPSVRV YADGETEDNS FSLKYRSNWT PGRFNSTGAK TKQWHYPSPY SRGALSVTSI DQGAYKRSGS SWGRPYEEK AGFGFSLDAR SCYSLFPVSQ NLTYIEVPQN VANRASTEVL QKVTQGNFNL GVALAEARST ASQLATQTIA L VKAYTAAR RGNWRQALRY LALNEDRKFR SKHVAGRWLE LQFGWLPLMS DIQGAYEMLT KVHLQEFLPM RAVRQVGTNI KL DGRLSYP AANFQTTCNI SRRIVIWFYI NDARLAWLSS LGILNPLGIV WEKVPFSFVV DWLLPVGNML EGLTAPVGCS YMS GTVTDV ITGESIISVD APYGWTVERQ GTAKAQISAM HRGVQSVWPT TGAYVKSPFS MVHTLDALAL IRQRLSR UniProtKB: Maturation protein A |
-Macromolecule #3: phage MS2 genome
| Macromolecule | Name: phage MS2 genome / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage MS2 (virus) Enterobacteria phage MS2 (virus) |
| Molecular weight | Theoretical: 1.1472007499999999 MDa |
| Sequence | String: GGGUGGGACC CCUUUCGGGG UCCUGCUCAA CUUCCUGUCG AGCUAAUGCC AUUUUUAAUG UCUUUAGCGA GACGCUACCA UGGCUAUCG CUGUAGGUAG CCGGAAUUCC AUUCCUAGGA GGUUUGACCU GUGCGAGCUU UUAGUACCCU UGAUAGGGAG A ACGAGACC ...String: GGGUGGGACC CCUUUCGGGG UCCUGCUCAA CUUCCUGUCG AGCUAAUGCC AUUUUUAAUG UCUUUAGCGA GACGCUACCA UGGCUAUCG CUGUAGGUAG CCGGAAUUCC AUUCCUAGGA GGUUUGACCU GUGCGAGCUU UUAGUACCCU UGAUAGGGAG A ACGAGACC UUCGUCCCCU CCGUUCGCGU UUACGCGGAC GGUGAGACUG AAGAUAACUC AUUCUCUUUA AAAUAUCGUU CG AACUGGA CUCCCGGUCG UUUUAACUCG ACUGGGGCCA AAACGAAACA GUGGCACUAC CCCUCUCCGU AUUCACGGGG GGC GUUAAG UGUCACAUCG AUAGAUCAAG GUGCCUACAA GCGAAGUGGG UCAUCGUGGG GUCGCCCGUA CGAGGAGAAA GCCG GUUUC GGCUUCUCCC UCGACGCACG CUCCUGCUAC AGCCUCUUCC CUGUAAGCCA AAACUUGACU UACAUCGAAG UGCCG CAGA ACGUUGCGAA CCGGGCGUCG ACCGAAGUCC UGCAAAAGGU CACCCAGGGU AAUUUUAACC UUGGUGUUGC UUUAGC AGA GGCCAGGUCG ACAGCCUCAC AACUCGCGAC GCAAACCAUU GCGCUCGUGA AGGCGUACAC UGCCGCUCGU CGCGGUA AU UGGCGCCAGG CGCUCCGCUA CCUUGCCCUA AACGAAGAUC GAAAGUUUCG AUCAAAACAC GUGGCCGGCA GGUGGUUG G AGUUGCAGUU CGGUUGGUUA CCACUAAUGA GUGAUAUCCA GGGUGCAUAU GAGAUGCUUA CGAAGGUUCA CCUUCAAGA GUUUCUUCCU AUGAGAGCCG UACGUCAGGU CGGUACUAAC AUCAAGUUAG AUGGCCGUCU GUCGUAUCCA GCUGCAAACU UCCAGACAA CGUGCAACAU AUCGCGACGU AUCGUGAUAU GGUUUUACAU AAACGAUGCA CGUUUGGCAU GGUUGUCGUC U CUAGGUAU CUUGAACCCA CUAGGUAUAG UGUGGGAAAA GGUGCCUUUC UCAUUCGUUG UCGACUGGCU CCUACCUGUA GG UAACAUG CUCGAGGGCC UUACGGCCCC CGUGGGAUGC UCCUACAUGU CAGGAACAGU UACUGACGUA AUAACGGGUG AGU CCAUCA UAAGCGUUGA CGCUCCCUAC GGGUGGACUG UGGAGAGACA GGGCACUGCU AAGGCCCAAA UCUCAGCCAU GCAU CGAGG GGUACAAUCC GUAUGGCCAA CAACUGGCGC GUACGUAAAG UCUCCUUUCU CGAUGGUCCA UACCUUAGAU GCGUU AGCA UUAAUCAGGC AACGGCUCUC UAGAUAGAGC CCUCAACCGG AGUUUGAAGC AUGGCUUCUA ACUUUACUCA GUUCGU UCU CGUCGACAAU GGCGGAACUG GCGACGUGAC UGUCGCCCCA AGCAACUUCG CUAACGGGGU CGCUGAAUGG AUCAGCU CU AACUCGCGUU CACAGGCUUA CAAAGUAACC UGUAGCGUUC GUCAGAGCUC UGCGCAGAAU CGCAAAUACA CCAUCAAA G UCGAGGUGCC UAAAGUGGCA ACCCAGACUG UUGGUGGUGU AGAGCUUCCU GUAGCCGCAU GGCGUUCGUA CUUAAAUAU GGAACUAACC AUUCCAAUUU UCGCUACGAA UUCCGACUGC GAGCUUAUUG UUAAGGCAAU GCAAGGUCUC CUAAAAGAUG GAAACCCGA UUCCCUCAGC AAUCGCAGCA AACUCCGGCA UCUACUAAUA GACGCCGGCC AUUCAAACAU GAGGAUUACC C AUGUCGAA GACAACAAAG AAGUUCAACU CUUUAUGUAU UGAUCUUCCU CGCGAUCUUU CUCUCGAAAU UUACCAAUCA AU UGCUUCU GUCGCUACUG GAAGCGGUGA UCCGCACAGU GACGACUUUA CAGCAAUUGC UUACUUAAGG GACGAAUUGC UCA CAAAGC AUCCGACCUU AGGUUCUGGU AAUGACGAGG CGACCCGUCG UACCUUAGCU AUCGCUAAGC UACGGGAGGC GAAU GGUGA UCGCGGUCAG AUAAAUAGAG AAGGUUUCUU ACAUGACAAA UCCUUGUCAU GGGAUCCGGA UGUUUUACAA ACCAG CAUC CGUAGCCUUA UUGGCAACCU CCUCUCUGGC UACCGAUCGU CGUUGUUUGG GCAAUGCACG UUCUCCAACG GUGCUC CUA UGGGGCACAA GUUGCAGGAU GCAGCGCCUU ACAAGAAGUU CGCUGAACAA GCAACCGUUA CCCCCCGCGC UCUGAGA GC GGCUCUAUUG GUCCGAGACC AAUGUGCGCC GUGGAUCAGA CACGCGGUCC GCUAUAACGA GUCAUAUGAA UUUAGGCU C GUUGUAGGGA ACGGAGUGUU UACAGUUCCG AAGAAUAAUA AAAUAGAUCG GGCUGCCUGU AAGGAGCCUG AUAUGAAUA UGUACCUCCA GAAAGGGGUC GGUGCUUUCA UCAGACGCCG GCUCAAAUCC GUUGGUAUAG ACCUGAAUGA UCAAUCGAUC AACCAGCGU CUGGCUCAGC AGGGCAGCGU AGAUGGUUCG CUUGCGACGA UAGACUUAUC GUCUGCAUCC GAUUCCAUCU C CGAUCGCC UGGUGUGGAG UUUUCUCCCA CCAGAGCUAU AUUCAUAUCU CGAUCGUAUC CGCUCACACU ACGGAAUCGU AG AUGGCGA GACGAUACGA UGGGAACUAU UUUCCACAAU GGGAAAUGGG UUCACAUUUG AGCUAGAGUC CAUGAUAUUC UGG GCAAUA GUCAAAGCGA CCCAAAUCCA UUUUGGUAAC GCCGGAACCA UAGGCAUCUA CGGGGACGAU AUUAUAUGUC CCAG UGAGA UUGCACCCCG UGUGCUAGAG GCACUUGCCU ACUACGGUUU UAAACCGAAU CUUCGUAAAA CGUUCGUGUC CGGGC UCUU UCGCGAGAGC UGCGGCGCGC ACUUUUACCG UGGUGUCGAU GUCAAACCGU UUUACAUCAA GAAACCUGUU GACAAU CUC UUCGCCCUGA UGCUGAUAUU AAAUCGGCUA CGGGGUUGGG GAGUUGUCGG AGGUAUGUCA GAUCCACGCC UCUAUAA GG UGUGGGUACG GCUCUCCUCC CAGGUGCCUU CGAUGUUCUU CGGUGGGACG GACCUCGCUG CCGACUACUA CGUAGUCA G CCCGCCUACG GCAGUCUCGG UAUACACCAA GACUCCGUAC GGGCGGCUGC UCGCGGAUAC CCGUACCUCG GGUUUCCGU CUUGCUCGUA UCGCUCGAGA ACGCAAGUUC UUCAGCGAAA AGCACGACAG UGGUCGCUAC AUAGCGUGGU UCCAUACUGG AGGUGAAAU CACCGACAGC AUGAAGUCCG CCGGCGUGCG CGUUAUACGC ACUUCGGAGU GGCUAACGCC GGUUCCCACA U UCCCUCAG GAGUGUGGGC CAGCGAGCUC UCCUCGGUAG CUGACCGAGG GACCCCCGUA AACGGGGUGG GUGUGCUCGA AA GAGCACG GGUGCGAAAG CGGUCCGGCU CCACCGAAAG GUGGGCGGGC UUCGGCCCAG GGACCUCCCC CUAAAGAGAG GAC CCGGGA UUCUCCCGAU UUGGUAACUA GCUGCUUGGC UAGUUACCAC CCA GENBANK: GENBANK: V00642.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: pH7.4 PBS |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.0 K |
| Specialist optics | Energy filter - Name: GIF / Energy filter - Lower energy threshold: 20 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | EFTEM mode with Gatan GIF energy filter. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Frames/image: 1-14 / Number grids imaged: 2 / Number real images: 6080 / Average exposure time: 5.8 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 47170 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-5tc1: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)