[English] 日本語
 Yorodumi
Yorodumi- EMDB-7631: Cryo-electron microscopy structure of infectious bronchitis coron... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7631 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
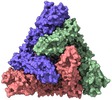
| Title | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | |||||||||
 Map data Map data | Infectious bronchitis coronavirus spike protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | infectious bronchitis coronavirus / spike / pre-fusion / cryo-EM / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Infectious bronchitis virus Infectious bronchitis virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.93 Å | |||||||||
 Authors Authors | Shang J / Zheng Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2018 Journal: PLoS Pathog / Year: 2018Title: Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. Authors: Jian Shang / Yuan Zheng / Yang Yang / Chang Liu / Qibin Geng / Chuming Luo / Wei Zhang / Fang Li /  Abstract: As cell-invading molecular machinery, coronavirus spike proteins pose an evolutionary conundrum due to their high divergence. In this study, we determined the cryo-EM structure of avian infectious ...As cell-invading molecular machinery, coronavirus spike proteins pose an evolutionary conundrum due to their high divergence. In this study, we determined the cryo-EM structure of avian infectious bronchitis coronavirus (IBV) spike protein from the γ-genus. The trimeric IBV spike ectodomain contains three receptor-binding S1 heads and a trimeric membrane-fusion S2 stalk. While IBV S2 is structurally similar to those from the other genera, IBV S1 possesses structural features that are unique to different other genera, thereby bridging these diverse spikes into an evolutionary spectrum. Specifically, among different genera, the two domains of S1, the N-terminal domain (S1-NTD) and C-terminal domain (S1-CTD), diverge from simpler tertiary structures and quaternary packing to more complex ones, leading to different functions of the spikes in receptor usage and membrane fusion. Based on the above structural and functional comparisons, we propose that the evolutionary spectrum of coronavirus spikes follows the order of α-, δ-, γ-, and β-genus. This study has provided insight into the evolutionary relationships among coronavirus spikes and deepened our understanding of their structural and functional diversity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7631.map.gz emd_7631.map.gz | 119.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7631-v30.xml emd-7631-v30.xml emd-7631.xml emd-7631.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7631.png emd_7631.png | 205.4 KB | ||
| Filedesc metadata |  emd-7631.cif.gz emd-7631.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7631 http://ftp.pdbj.org/pub/emdb/structures/EMD-7631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7631 | HTTPS FTP |
-Related structure data
| Related structure data |  6cv0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7631.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7631.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Infectious bronchitis coronavirus spike protein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
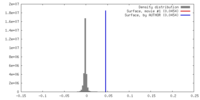
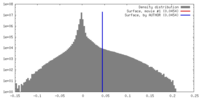
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : homotrimer of infectious bronchitis coronavirus spike ectodomain
| Entire | Name: homotrimer of infectious bronchitis coronavirus spike ectodomain |
|---|---|
| Components |
|
-Supramolecule #1: homotrimer of infectious bronchitis coronavirus spike ectodomain
| Supramolecule | Name: homotrimer of infectious bronchitis coronavirus spike ectodomain type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Infectious bronchitis virus Infectious bronchitis virus |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Infectious bronchitis virus Infectious bronchitis virus |
| Molecular weight | Theoretical: 121.957703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ALYDSSSYVY YYQSAFRPPN GWHLHGGAYA VVNISSESNN AGSSPGCIVG TIHGGRVVNA SSIAMTAPSS GMAWSSSQFC TAHCNFSDT TVFVTHCYKY DGCPITGMLQ KNFLRVSAMK NGQLFYNLTV SVAKYPTFKS FQCVNNLTSV YLNGDLVYTS N ETTDVTSA ...String: ALYDSSSYVY YYQSAFRPPN GWHLHGGAYA VVNISSESNN AGSSPGCIVG TIHGGRVVNA SSIAMTAPSS GMAWSSSQFC TAHCNFSDT TVFVTHCYKY DGCPITGMLQ KNFLRVSAMK NGQLFYNLTV SVAKYPTFKS FQCVNNLTSV YLNGDLVYTS N ETTDVTSA GVYFKAGGPI TYKVMREVKA LAYFVNGTAQ DVILCDGSPR GLLACQYNTG NFSDGFYPFI NSSLVKQKFI VY RENSVNT TFTLHNFTFH NETGANPNPS GVQNIQTYQT QTAQSGYYNF NFSFLSSFVY KESNFMYGSY HPSCNFRLET INN GLWFNS LSVSIAYGPL QGGCKQSVFS GRATCCYAYS YGGPSLCKGV YSGELDLNFE CGLLVYVTKS GGSRIQTATE PPVI TRHNY NNITLNTCVD YNIYGRTGQG FITNVTDSAV SYNYLADAGL AILDTSGSID IFVVQGEYGL TYYKVNPCED VNQQF VVSG GKLVGILTSR NETGSQLLEN QFYIKITNGT RRFRRSITEN VANCPYVSYG KFCIKPDGSI ATIVPKQLEQ FVAPLL NVT ENVLIPNSFN LTVTDEYIQT RMDKVQINCL QYVCGNSLDC RDLFQQYGPV CDNILSVVNS IGQKEDMELL NFYSSTK PA GFNTPFLSNV STGEFNISLL LTTPSSPRRR SFIEDLLFTS VESVGLPTDD AYKNCTAGPL GFLKDLACAR EYNGLLVL P PIITAEMQTL YTSSLVASMA FGGITAAGAI PFATQLQARI NHLGITQSLL LKNQEKIAAS FNKAIGRMQE GFRSTSLAL QQIQHVVNKQ NAILTETMAS LNKNFGAISS LIQEIYQQLD AIQANAQVDR LITGRLSSLS VLASAKQAEH IRVSQQRELA TQKINECVK SQSIRYSFCG NGRHVLTIPQ NAPNGIVFIH FSYTPDSFVN VTAIVGFCVK PANASQYAIV PANGRGIFIQ V NGSYYITA RDMYMPRAIT AGDIVTLTSC QANYVSVNKT VITTFVDNDD FDFNDELSKW WNDTKHELPD FDKFNYTVPI LD IDSEIDR IQGVIQGLND SVDIKQIEDK IEEILSKIYH IENEIARIKK LIGEIGGGGS HHHHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 39 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 5.366 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)