+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7348 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast Vacuolar ATPase Vo in lipid nanodisc | |||||||||||||||
 Map data Map data | The CryoEM Structure of Nanodisc Reconstituted Yeast Vacuolar ATPase Vo Proton Channel | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Vacuolar H+-ATPase / Vo proton channel / rotary motor enzyme / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcell wall mannoprotein biosynthetic process / ATPase-coupled ion transmembrane transporter activity / cellular response to alkaline pH / protein localization to vacuolar membrane / Insulin receptor recycling / Transferrin endocytosis and recycling / polyphosphate metabolic process / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / Golgi lumen acidification ...cell wall mannoprotein biosynthetic process / ATPase-coupled ion transmembrane transporter activity / cellular response to alkaline pH / protein localization to vacuolar membrane / Insulin receptor recycling / Transferrin endocytosis and recycling / polyphosphate metabolic process / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / Golgi lumen acidification / P-type proton-exporting transporter activity / vacuolar transport / vacuolar proton-transporting V-type ATPase, V0 domain / endosomal lumen acidification / vacuole organization / protein targeting to vacuole / proton-transporting V-type ATPase complex / fungal-type vacuole / vacuolar proton-transporting V-type ATPase complex / cellular hyperosmotic response / vacuolar acidification / fungal-type vacuole membrane / phosphatidylinositol-3,5-bisphosphate binding / proton transmembrane transporter activity / proton-transporting ATPase activity, rotational mechanism / intracellular copper ion homeostasis / Neutrophil degranulation / RNA endonuclease activity / proton transmembrane transport / cell periphery / transmembrane transport / endocytosis / ATPase binding / protein-containing complex assembly / intracellular iron ion homeostasis / membrane raft / Golgi membrane / endoplasmic reticulum membrane / membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
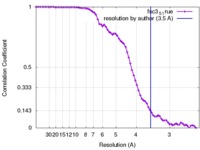
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Roh S / Stam NJ | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2018 Journal: Mol Cell / Year: 2018Title: The 3.5-Å CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase V Proton Channel. Authors: Soung-Hun Roh / Nicholas J Stam / Corey F Hryc / Sergio Couoh-Cardel / Grigore Pintilie / Wah Chiu / Stephan Wilkens /  Abstract: The molecular mechanism of transmembrane proton translocation in rotary motor ATPases is not fully understood. Here, we report the 3.5-Å resolution cryoEM structure of the lipid nanodisc- ...The molecular mechanism of transmembrane proton translocation in rotary motor ATPases is not fully understood. Here, we report the 3.5-Å resolution cryoEM structure of the lipid nanodisc-reconstituted V proton channel of the yeast vacuolar H-ATPase, captured in a physiologically relevant, autoinhibited state. The resulting atomic model provides structural detail for the amino acids that constitute the proton pathway at the interface of the proteolipid ring and subunit a. Based on the structure and previous mutagenesis studies, we propose the chemical basis of transmembrane proton transport. Moreover, we discovered that the C terminus of the assembly factor Voa1 is an integral component of mature V. Voa1's C-terminal transmembrane α helix is bound inside the proteolipid ring, where it contributes to the stability of the complex. Our structure rationalizes possible mechanisms by which mutations in human V can result in disease phenotypes and may thus provide new avenues for therapeutic interventions. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7348.map.gz emd_7348.map.gz | 28.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7348-v30.xml emd-7348-v30.xml emd-7348.xml emd-7348.xml | 30.5 KB 30.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7348_fsc.xml emd_7348_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_7348.png emd_7348.png | 338.3 KB | ||
| Filedesc metadata |  emd-7348.cif.gz emd-7348.cif.gz | 7.9 KB | ||
| Others |  emd_7348_additional.map.gz emd_7348_additional.map.gz emd_7348_half_map_1.map.gz emd_7348_half_map_1.map.gz emd_7348_half_map_2.map.gz emd_7348_half_map_2.map.gz | 28.6 MB 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7348 http://ftp.pdbj.org/pub/emdb/structures/EMD-7348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7348 | HTTPS FTP |
-Related structure data
| Related structure data |  6c6lMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7348.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7348.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
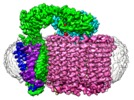
| Annotation | The CryoEM Structure of Nanodisc Reconstituted Yeast Vacuolar ATPase Vo Proton Channel | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
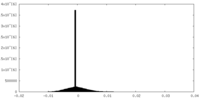
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_7348_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
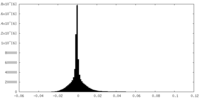
| Density Histograms |
-Half map: #1
| File | emd_7348_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
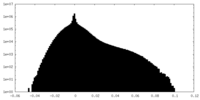
| Density Histograms |
-Half map: #2
| File | emd_7348_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
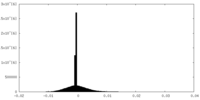
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast Vacuolar ATPase Vo
| Entire | Name: Yeast Vacuolar ATPase Vo |
|---|---|
| Components |
|
-Supramolecule #1: Yeast Vacuolar ATPase Vo
| Supramolecule | Name: Yeast Vacuolar ATPase Vo / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: V-type proton ATPase subunit c'
| Macromolecule | Name: V-type proton ATPase subunit c' / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 17.046361 KDa |
| Sequence | String: MSTQLASNIY APLYAPFFGF AGCAAAMVLS CLGAAIGTAK SGIGIAGIGT FKPELIMKSL IPVVMSGILA IYGLVVAVLI AGNLSPTED YTLFNGFMHL SCGLCVGFAC LSSGYAIGMV GDVGVRKYMH QPRLFVGIVL ILIFSEVLGL YGMIVALILN T RGSE UniProtKB: V-type proton ATPase subunit c' |
-Macromolecule #2: V-type proton ATPase subunit c''
| Macromolecule | Name: V-type proton ATPase subunit c'' / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 22.610641 KDa |
| Sequence | String: MNKESKDDDM SLGKFSFSHF LYYLVLIVVI VYGLYKLFTG HGSDINFGKF LLRTSPYMWA NLGIALCVGL SVVGAAWGIF ITGSSMIGA GVRAPRITTK NLISIIFCEV VAIYGLIIAI VFSSKLTVAT AENMYSKSNL YTGYSLFWAG ITVGASNLIC G IAVGITGA ...String: MNKESKDDDM SLGKFSFSHF LYYLVLIVVI VYGLYKLFTG HGSDINFGKF LLRTSPYMWA NLGIALCVGL SVVGAAWGIF ITGSSMIGA GVRAPRITTK NLISIIFCEV VAIYGLIIAI VFSSKLTVAT AENMYSKSNL YTGYSLFWAG ITVGASNLIC G IAVGITGA TAAISDAADS ALFVKILVIE IFGSILGLLG LIVGLLMAGK ASEFQ UniProtKB: V-type proton ATPase subunit c'' |
-Macromolecule #3: V0 assembly protein 1
| Macromolecule | Name: V0 assembly protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 29.694885 KDa |
| Sequence | String: MVFGQLYALF IFTLSCCISK TVQADSSKES SSFISFDKES NWDTISTISS TADVISSVDS AIAVFEFDNF SLLDNLMIDE EYPFFNRFF ANDVSLTVHD DSPLNISQSL SPIMEQFTVD ELPESASDLL YEYSLDDKSI VLFKFTSDAY DLKKLDEFID S CLSFLEDK ...String: MVFGQLYALF IFTLSCCISK TVQADSSKES SSFISFDKES NWDTISTISS TADVISSVDS AIAVFEFDNF SLLDNLMIDE EYPFFNRFF ANDVSLTVHD DSPLNISQSL SPIMEQFTVD ELPESASDLL YEYSLDDKSI VLFKFTSDAY DLKKLDEFID S CLSFLEDK SGDNLTVVIN SLGWAFEDED GDDEYATEET LSHHDNNKGK EGDDDILSSI WTEGLLMCLI VSALLLFILI VA LSWISNL DITYGALEKS TNPIKKNN UniProtKB: V0 assembly protein 1 |
-Macromolecule #4: V-type proton ATPase subunit e
| Macromolecule | Name: V-type proton ATPase subunit e / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 8.387065 KDa |
| Sequence | String: MSSFYTVVGV FIVVSAMSVL FWIMAPKNNQ AVWRSTVILT LAMMFLMWAI TFLCQLHPLV APRRSDLRPE FAE UniProtKB: V-type proton ATPase subunit e |
-Macromolecule #5: V-type proton ATPase subunit c
| Macromolecule | Name: V-type proton ATPase subunit c / type: protein_or_peptide / ID: 5 / Number of copies: 8 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 16.357501 KDa |
| Sequence | String: MTELCPVYAP FFGAIGCASA IIFTSLGAAY GTAKSGVGIC ATCVLRPDLL FKNIVPVIMA GIIAIYGLVV SVLVCYSLGQ KQALYTGFI QLGAGLSVGL SGLAAGFAIG IVGDAGVRGS SQQPRLFVGM ILILIFAEVL GLYGLIVALL LNSRATQDVV C UniProtKB: V-type proton ATPase subunit c |
-Macromolecule #6: V-type proton ATPase subunit d
| Macromolecule | Name: V-type proton ATPase subunit d / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 39.822484 KDa |
| Sequence | String: MEGVYFNIDN GFIEGVVRGY RNGLLSNNQY INLTQCDTLE DLKLQLSSTD YGNFLSSVSS ESLTTSLIQE YASSKLYHEF NYIRDQSSG STRKFMDYIT YGYMIDNVAL MITGTIHDRD KGEILQRCHP LGWFDTLPTL SVATDLESLY ETVLVDTPLA P YFKNCFDT ...String: MEGVYFNIDN GFIEGVVRGY RNGLLSNNQY INLTQCDTLE DLKLQLSSTD YGNFLSSVSS ESLTTSLIQE YASSKLYHEF NYIRDQSSG STRKFMDYIT YGYMIDNVAL MITGTIHDRD KGEILQRCHP LGWFDTLPTL SVATDLESLY ETVLVDTPLA P YFKNCFDT AEELDDMNIE IIRNKLYKAY LEDFYNFVTE EIPEPAKECM QTLLGFEADR RSINIALNSL QSSDIDPDLK SD LLPNIGK LYPLATFHLA QAQDFEGVRA ALANVYEYRG FLETGNLEDH FYQLEMELCR DAFTQQFAIS TVWAWMKSKE QEV RNITWI AECIAQNQRE RINNYISVY UniProtKB: V-type proton ATPase subunit d |
-Macromolecule #7: V-type proton ATPase subunit a, vacuolar isoform
| Macromolecule | Name: V-type proton ATPase subunit a, vacuolar isoform / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 95.625484 KDa |
| Sequence | String: MAEKEEAIFR SAEMALVQFY IPQEISRDSA YTLGQLGLVQ FRDLNSKVRA FQRTFVNEIR RLDNVERQYR YFYSLLKKHD IKLYEGDTD KYLDGSGELY VPPSGSVIDD YVRNASYLEE RLIQMEDATD QIEVQKNDLE QYRFILQSGD EFFLKGDNTD S TSYMDEDM ...String: MAEKEEAIFR SAEMALVQFY IPQEISRDSA YTLGQLGLVQ FRDLNSKVRA FQRTFVNEIR RLDNVERQYR YFYSLLKKHD IKLYEGDTD KYLDGSGELY VPPSGSVIDD YVRNASYLEE RLIQMEDATD QIEVQKNDLE QYRFILQSGD EFFLKGDNTD S TSYMDEDM IDANGENIAA AIGASVNYVT GVIARDKVAT LEQILWRVLR GNLFFKTVEI EQPVYDVKTR EYKHKNAFIV FS HGDLIIK RIRKIAESLD ANLYDVDSSN EGRSQQLAKV NKNLSDLYTV LKTTSTTLES ELYAIAKELD SWFQDVTREK AIF EILNKS NYDTNRKILI AEGWIPRDEL ATLQARLGEM IARLGIDVPS IIQVLDTNHT PPTFHRTNKF TAGFQSICDC YGIA QYREI NAGLPTIVTF PFMFAIMFGD MGHGFLMTLA ALSLVLNEKK INKMKRGEIF DMAFTGRYII LLMGVFSMYT GFLYN DIFS KTMTIFKSGW KWPDHWKKGE SITATSVGTY PIGLDWAWHG TENALLFSNS YKMKLSILMG FIHMTYSYFF SLANHL YFN SMIDIIGNFI PGLLFMQGIF GYLSVCIVYK WAVDWVKDGK PAPGLLNMLI NMFLSPGTID DELYPHQAKV QVFLLLM AL VCIPWLLLVK PLHFKFTHKK KSHEPLPSTE ADASSEDLEA QQLISAMDAD DAEEEEVGSG SHGEDFGDIM IHQVIHTI E FCLNCVSHTA SYLRLWALSL AHAQLSSVLW TMTIQIAFGF RGFVGVFMTV ALFAMWFALT CAVLVLMEGT SAMLHSLRL HWVESMSKFF VGEGLPYEPF AFEYKDMEVA VASASSSASS UniProtKB: V-type proton ATPase subunit a, vacuolar isoform |
-Macromolecule #8: V-type proton ATPase subunit f
| Macromolecule | Name: V-type proton ATPase subunit f / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 9.369934 KDa |
| Sequence | String: MRPVVSTGKA WCCTVLSAFG VVILSVIAHL FNTNHESFVG SINDPEDGPA VAHTVYLAAL VYLVFFVFCG FQVYLARRKP SIELR UniProtKB: V-type proton ATPase subunit f |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Sugar embedding | Material: ice |
| Grid | Model: Quantifoil, UltrAuFoil / Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)