[English] 日本語
 Yorodumi
Yorodumi- EMDB-7305: The Structure of Full-Length Kv Beta 2.1 Determined by Cryogenic ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7305 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Structure of Full-Length Kv Beta 2.1 Determined by Cryogenic Electron Microscopy | |||||||||
 Map data Map data | Raw unsharpened reconstruction of Kv Beta 2.1. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpinceau fiber / methylglyoxal reductase (NADPH) (acetol producing) activity / Voltage gated Potassium channels / potassium channel complex / regulation of protein localization to cell surface / : / axon initial segment / Oxidoreductases; Acting on the CH-OH group of donors; With NAD+ or NADP+ as acceptor / juxtaparanode region of axon / myoblast differentiation ...pinceau fiber / methylglyoxal reductase (NADPH) (acetol producing) activity / Voltage gated Potassium channels / potassium channel complex / regulation of protein localization to cell surface / : / axon initial segment / Oxidoreductases; Acting on the CH-OH group of donors; With NAD+ or NADP+ as acceptor / juxtaparanode region of axon / myoblast differentiation / regulation of potassium ion transmembrane transport / Neutrophil degranulation / neuromuscular process / voltage-gated potassium channel activity / potassium channel regulator activity / hematopoietic progenitor cell differentiation / voltage-gated potassium channel complex / axon terminus / postsynaptic density membrane / cytoplasmic side of plasma membrane / transmembrane transporter binding / cytoskeleton / neuron projection / postsynaptic density / axon / protein-containing complex binding / glutamatergic synapse / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
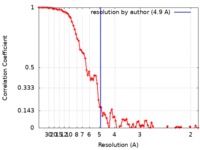
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Spear JM / Mendez JH / Stagg SM | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The Structure of Full-Length Kv Beta 2.1 Determined by Cryogenic Electron Microscopy Authors: Spear JM / Mendez JH / Danyuk Y / Stagg SM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7305.map.gz emd_7305.map.gz | 36.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7305-v30.xml emd-7305-v30.xml emd-7305.xml emd-7305.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7305_fsc.xml emd_7305_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_7305.png emd_7305.png | 84.7 KB | ||
| Others |  emd_7305_additional_1.map.gz emd_7305_additional_1.map.gz emd_7305_additional_2.map.gz emd_7305_additional_2.map.gz emd_7305_half_map_1.map.gz emd_7305_half_map_1.map.gz emd_7305_half_map_2.map.gz emd_7305_half_map_2.map.gz | 47.9 MB 49.1 MB 4 MB 4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7305 http://ftp.pdbj.org/pub/emdb/structures/EMD-7305 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7305 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7305 | HTTPS FTP |
-Related structure data
| Related structure data |  6ci1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7305.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7305.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw unsharpened reconstruction of Kv Beta 2.1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.953 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
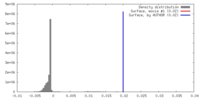
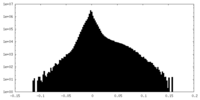
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: None
| File | emd_7305_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
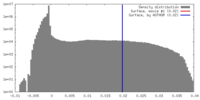
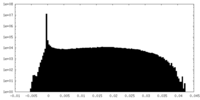
| Density Histograms |
-Additional map: None
| File | emd_7305_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
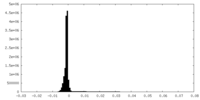
| Density Histograms |
-Half map: Independently masked half map
| File | emd_7305_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Independently masked half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
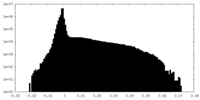
| Density Histograms |
-Half map: Independently masked half map
| File | emd_7305_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Independently masked half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octamer of Kv Beta 2.1
| Entire | Name: Octamer of Kv Beta 2.1 |
|---|---|
| Components |
|
-Supramolecule #1: Octamer of Kv Beta 2.1
| Supramolecule | Name: Octamer of Kv Beta 2.1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Kv Beta
| Macromolecule | Name: Kv Beta / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MYPESTTGSP ARLSLRQTGS PGMIYSTRYG SPKRQLQFYR NLGKSGLRVS CLGLGTWVTF GGQITDEMAE HLMTLAYDN GINLFDTAEV YAAGKAEVVL GNIIKKKGWR RSSLVITTKI FWGGKAETER GLSRKHIIEG L KASLERLQ LEYVDVVFAN RPDPNTPMEE ...String: MYPESTTGSP ARLSLRQTGS PGMIYSTRYG SPKRQLQFYR NLGKSGLRVS CLGLGTWVTF GGQITDEMAE HLMTLAYDN GINLFDTAEV YAAGKAEVVL GNIIKKKGWR RSSLVITTKI FWGGKAETER GLSRKHIIEG L KASLERLQ LEYVDVVFAN RPDPNTPMEE TVRAMTHVIN QGMAMYWGTS RWSSMEIMEA YSVARQFNLI PP ICEQAEY HMFQREKVEV QLPELFHKIG VGAMTWSPLA CGIVSGKYDS GIPPYSRASL KGYQWLKDKI LSE EGRRQQ AKLKELQAIA ERLGCTLPQL AIAWCLRNEG VSSVLLGASN AEQLMENIGA IQVLPKLSSS IVHE IDSIL GNKPYSKKDY RS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6ci1: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)