[English] 日本語
 Yorodumi
Yorodumi- EMDB-7293: Cryo-EM structure of CENP-A nucleosome in complex with kinetochor... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7293 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
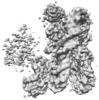
| Title | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | |||||||||
 Map data Map data | Cryo-EM density map of CENP-A nucleosome in complex with kinetochore protein CENP-N | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | STRUCTURAL PROTEIN-DNA complex / Histone fold / Centromeric nucleosome / Kinetochore | |||||||||
| Function / homology |  Function and homology information Function and homology informationCENP-A containing chromatin assembly / protein localization to chromosome, centromeric region / kinetochore assembly / condensed chromosome, centromeric region / inner kinetochore / mitotic cytokinesis / establishment of mitotic spindle orientation / carbohydrate transmembrane transporter activity / chromosome, centromeric region / maltose binding ...CENP-A containing chromatin assembly / protein localization to chromosome, centromeric region / kinetochore assembly / condensed chromosome, centromeric region / inner kinetochore / mitotic cytokinesis / establishment of mitotic spindle orientation / carbohydrate transmembrane transporter activity / chromosome, centromeric region / maltose binding / maltose transport / maltodextrin transmembrane transport / negative regulation of tumor necrosis factor-mediated signaling pathway / pericentric heterochromatin / negative regulation of megakaryocyte differentiation / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / protein localization to CENP-A containing chromatin / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / Mitotic Prometaphase / telomere organization / EML4 and NUDC in mitotic spindle formation / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / RNA Polymerase I Promoter Opening / Inhibition of DNA recombination at telomere / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / Resolution of Sister Chromatid Cohesion / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / innate immune response in mucosa / Defective pyroptosis / Negative Regulation of CDH1 Gene Transcription / HDACs deacetylate histones / chromosome segregation / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / RNA Polymerase I Promoter Escape / Nonhomologous End-Joining (NHEJ) / lipopolysaccharide binding / Transcriptional regulation by small RNAs / RHO GTPases Activate Formins / Formation of the beta-catenin:TCF transactivating complex / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / HDMs demethylate histones / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / DNA Damage/Telomere Stress Induced Senescence / Pre-NOTCH Transcription and Translation / Meiotic recombination / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Metalloprotease DUBs / Transcriptional regulation of granulopoiesis / RMTs methylate histone arginines / HCMV Early Events / structural constituent of chromatin / Separation of Sister Chromatids / UCH proteinases / heterochromatin formation / nucleosome / antimicrobial humoral immune response mediated by antimicrobial peptide / nucleosome assembly / antibacterial humoral response / E3 ubiquitin ligases ubiquitinate target proteins / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / outer membrane-bounded periplasmic space / HATs acetylate histones / RUNX1 regulates transcription of genes involved in differentiation of HSCs / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / chromatin organization / Processing of DNA double-strand break ends / Senescence-Associated Secretory Phenotype (SASP) / Oxidative Stress Induced Senescence / Estrogen-dependent gene expression / killing of cells of another organism / defense response to Gram-negative bacterium / chromosome, telomeric region / Ub-specific processing proteases / defense response to Gram-positive bacterium / protein heterodimerization activity / Amyloid fiber formation / negative regulation of cell population proliferation / chromatin binding / protein-containing complex Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.92 Å | |||||||||
 Authors Authors | Chittori S / Hong J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Authors: Sagar Chittori / Jingjun Hong / Hayden Saunders / Hanqiao Feng / Rodolfo Ghirlando / Alexander E Kelly / Yawen Bai / Sriram Subramaniam /  Abstract: Accurate chromosome segregation requires the proper assembly of kinetochore proteins. A key step in this process is the recognition of the histone H3 variant CENP-A in the centromeric nucleosome by ...Accurate chromosome segregation requires the proper assembly of kinetochore proteins. A key step in this process is the recognition of the histone H3 variant CENP-A in the centromeric nucleosome by the kinetochore protein CENP-N. We report cryo-electron microscopy (cryo-EM), biophysical, biochemical, and cell biological studies of the interaction between the CENP-A nucleosome and CENP-N. We show that human CENP-N confers binding specificity through interactions with the L1 loop of CENP-A, stabilized by electrostatic interactions with the nucleosomal DNA. Mutational analyses demonstrate analogous interactions in , which are further supported by residue-swapping experiments involving the L1 loop of CENP-A. Our results are consistent with the coevolution of CENP-N and CENP-A and establish the structural basis for recognition of the CENP-A nucleosome to enable kinetochore assembly and centromeric chromatin organization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7293.map.gz emd_7293.map.gz | 537.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7293-v30.xml emd-7293-v30.xml emd-7293.xml emd-7293.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7293.png emd_7293.png | 88.1 KB | ||
| Filedesc metadata |  emd-7293.cif.gz emd-7293.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7293 http://ftp.pdbj.org/pub/emdb/structures/EMD-7293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7293 | HTTPS FTP |
-Related structure data
| Related structure data |  6buzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7293.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7293.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM density map of CENP-A nucleosome in complex with kinetochore protein CENP-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Centromeric nucleosome in complex with kinetochore protein CENP-N
| Entire | Name: Centromeric nucleosome in complex with kinetochore protein CENP-N |
|---|---|
| Components |
|
-Supramolecule #1: Centromeric nucleosome in complex with kinetochore protein CENP-N
| Supramolecule | Name: Centromeric nucleosome in complex with kinetochore protein CENP-N type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Histone H3-like centromeric protein A
| Macromolecule | Name: Histone H3-like centromeric protein A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.195002 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MGPRRRSRKP EAPRRRSPSP TPTPGPSRRG PSLGASSHQH SRRRQGWLKE IRKLQKSTHL LIRKLPFSR LAREICVKFT RGVDFNWQAQ ALLALQEAAE AFLVHLFEDA YLLTLHAGRV TLFPKDVQLA RRIRGLEEGL G UniProtKB: Histone H3-like centromeric protein A |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.165551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VHRLLRKGNY SERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG RVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A type 1-B/E |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.935239 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPEPAKSAPA PKKGSKKAVT KAQKKDGKKR KRSRKESYSI YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV TKYTSAK UniProtKB: Histone H2B type 1-J |
-Macromolecule #7: Maltose-binding periplasmic protein, Centromere protein N chimera
| Macromolecule | Name: Maltose-binding periplasmic protein, Centromere protein N chimera type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.607797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKEHHHHHHH HAKIEEGKLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE HPDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLA EITPDKAFQD KLYPFTWDAV RYNGKLIAYP IAVEALSLIY NKDLLPNPPK TWEEIPALDK ELKAKGKSAL M FNLQEPYF ...String: MKEHHHHHHH HAKIEEGKLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE HPDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLA EITPDKAFQD KLYPFTWDAV RYNGKLIAYP IAVEALSLIY NKDLLPNPPK TWEEIPALDK ELKAKGKSAL M FNLQEPYF TWPLIAADGG YAFKYENGKY DIKDVGVDNA GAKAGLTFLV DLIKNKHMNA DTDYSIAEAA FNKGETAMTI NG PWAWSNI DTSKVNYGVT VLPTFKGQPS KPFVGVLSAG INAASPNKEL AKEFLENYLL TDEGLEAVNK DKPLGAVALK SYE EELAKD PRIAATMENA QKGEIMPNIP QMSAFWYAVR TAVINAASGR QTVDAALAAA QTNAAAMDET VAEFIKRTIL KIPM NELTT ILKAWDFLSE NQLQTVNFRQ RKESVVQHLI HLCEEKRASI SDAALLDIIY MQFHQHQKVW EVFQMSKGPG EDVDL FDMK QFKNSFKKIL QRALKNVTVS FRETEENAVW IRIAWGTQYT KPNQYKPTYV VYYSQTPYAF TSSSMLRRNT PLLGQA LTI ASKHHQIVKM DLRSRYLDSL KAIVFKQYNQ TFETHNSTTP LQERSLGLDI NMDSRIIHEN IVEKERVQRI TQETFGD YP QPQLEFAQYK LETKFKSGLN GSILAE UniProtKB: Maltose/maltodextrin-binding periplasmic protein, Centromere protein N |
-Macromolecule #5: DNA (147-MER)
| Macromolecule | Name: DNA (147-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.13877 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DG)(DA)(DT) |
-Macromolecule #6: DNA (147-MER)
| Macromolecule | Name: DNA (147-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.610043 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.92 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2.0) / Number images used: 61749 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
-Atomic model buiding 1
| Details | Ab initio modeling of CENP-N backbone trace |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Target criteria: Model to Map correlation, Molprobity |
| Output model |  PDB-6buz: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















