+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ymy | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cytochrome c oxidase from Saccharomyces cerevisiae | |||||||||||||||
 Components Components | (Cytochrome c oxidase subunit ...) x 12 | |||||||||||||||
 Keywords Keywords | ELECTRON TRANSPORT / CIV / CytcO | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial respiratory chain complex IV assembly / mitochondrial respirasome assembly / Mitochondrial protein degradation / cellular respiration / respiratory chain complex IV / respiratory chain complex / cytochrome-c oxidase / mitochondrial electron transport, cytochrome c to oxygen / cytochrome-c oxidase activity / ubiquinone binding ...mitochondrial respiratory chain complex IV assembly / mitochondrial respirasome assembly / Mitochondrial protein degradation / cellular respiration / respiratory chain complex IV / respiratory chain complex / cytochrome-c oxidase / mitochondrial electron transport, cytochrome c to oxygen / cytochrome-c oxidase activity / ubiquinone binding / ATP synthesis coupled electron transport / enzyme regulator activity / proton transmembrane transport / aerobic respiration / respiratory electron transport chain / mitochondrial intermembrane space / mitochondrial membrane / oxidoreductase activity / mitochondrial inner membrane / copper ion binding / heme binding / mitochondrion / zinc ion binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||||||||
 Authors Authors | Berndtsson, J. / Rathore, S. / Ott, M. | |||||||||||||||
| Funding support |  Sweden, 4items Sweden, 4items
| |||||||||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2020 Journal: EMBO Rep / Year: 2020Title: Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance. Authors: Jens Berndtsson / Andreas Kohler / Sorbhi Rathore / Lorena Marin-Buera / Hannah Dawitz / Jutta Diessl / Verena Kohler / Antoni Barrientos / Sabrina Büttner / Flavia Fontanesi / Martin Ott /    Abstract: Respiratory chains are crucial for cellular energy conversion and consist of multi-subunit complexes that can assemble into supercomplexes. These structures have been intensively characterized in ...Respiratory chains are crucial for cellular energy conversion and consist of multi-subunit complexes that can assemble into supercomplexes. These structures have been intensively characterized in various organisms, but their physiological roles remain unclear. Here, we elucidate their function by leveraging a high-resolution structural model of yeast respiratory supercomplexes that allowed us to inhibit supercomplex formation by mutation of key residues in the interaction interface. Analyses of a mutant defective in supercomplex formation, which still contains fully functional individual complexes, show that the lack of supercomplex assembly delays the diffusion of cytochrome c between the separated complexes, thus reducing electron transfer efficiency. Consequently, competitive cellular fitness is severely reduced in the absence of supercomplex formation and can be restored by overexpression of cytochrome c. In sum, our results establish how respiratory supercomplexes increase the efficiency of cellular energy conversion, thereby providing an evolutionary advantage for aerobic organisms. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ymy.cif.gz 6ymy.cif.gz | 342.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ymy.ent.gz pdb6ymy.ent.gz | 269.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ymy.json.gz 6ymy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ym/6ymy https://data.pdbj.org/pub/pdb/validation_reports/ym/6ymy ftp://data.pdbj.org/pub/pdb/validation_reports/ym/6ymy ftp://data.pdbj.org/pub/pdb/validation_reports/ym/6ymy | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  10848MC  6ymxC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Cytochrome c oxidase subunit ... , 12 types, 12 molecules abcdefghijkm
| #1: Protein | Mass: 58316.938 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P00401, cytochrome-c oxidase |
|---|---|
| #2: Protein | Mass: 26779.816 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P00410, cytochrome-c oxidase |
| #3: Protein | Mass: 30252.391 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P00420, cytochrome-c oxidase |
| #4: Protein | Mass: 12694.382 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P04037 |
| #5: Protein | Mass: 14324.147 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P00424 |
| #6: Protein | Mass: 11725.173 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P00427 |
| #7: Protein | Mass: 6410.639 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P10174 |
| #8: Protein | Mass: 5737.735 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P04039 |
| #9: Protein | Mass: 6056.131 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P07255 |
| #10: Protein | Mass: 9225.291 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: Q01519 |
| #11: Protein | Mass: 13306.061 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: P32799 |
| #12: Protein/peptide | Mass: 4347.023 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  References: UniProt: Q2V2P9 |
-Non-polymers , 7 types, 16 molecules 


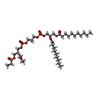









| #13: Chemical | ChemComp-CU / | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #14: Chemical | | #15: Chemical | ChemComp-PTY / #16: Chemical | ChemComp-CN3 / ( | #17: Chemical | ChemComp-CUA / | #18: Chemical | #19: Chemical | ChemComp-ZN / | |
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cytochrome c oxidase / Type: COMPLEX / Entity ID: #1-#12 / Source: NATURAL | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:  Strain: W303-1b / Organelle: Mitochondria | |||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: Homemade | |||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Nominal defocus max: -3 nm / Nominal defocus min: -1.4 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE |
| Specimen holder | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 9 sec. / Electron dose: 40 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 8775 |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
| Image scans | Width: 3838 / Height: 3710 / Movie frames/image: 40 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 647805 | ||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.41 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 201223 / Num. of class averages: 1 / Symmetry type: POINT |
 Movie
Movie Controller
Controller















 PDBj
PDBj









