[English] 日本語
 Yorodumi
Yorodumi- PDB-6s8m: S. pombe microtubule decorated with Cut7 motor domain in the AMPP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6s8m | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | MOTOR PROTEIN / S. pombe / microtubule / kinesin-5 / Cut7 motor domain | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitotic spindle pole body duplication / Platelet degranulation / mitotic spindle formation (spindle phase one) / mitotic spindle elongation (spindle phase three) / Kinesins / initial mitotic spindle pole body separation / microtubule plus-end directed mitotic chromosome migration / nuclear migration by microtubule mediated pushing forces / meiotic spindle pole / meiotic spindle assembly ...mitotic spindle pole body duplication / Platelet degranulation / mitotic spindle formation (spindle phase one) / mitotic spindle elongation (spindle phase three) / Kinesins / initial mitotic spindle pole body separation / microtubule plus-end directed mitotic chromosome migration / nuclear migration by microtubule mediated pushing forces / meiotic spindle pole / meiotic spindle assembly / nuclear division / mitotic spindle midzone / mitotic spindle pole body / mitotic spindle elongation / mitotic spindle midzone assembly / spindle elongation / polar microtubule / astral microtubule / plus-end-directed microtubule motor activity / meiotic spindle / microtubule motor activity / minus-end-directed microtubule motor activity / microtubule associated complex / intracellular distribution of mitochondria / mitotic spindle assembly / cytoplasmic microtubule / cytoplasmic microtubule organization / spindle microtubule / kinetochore / structural constituent of cytoskeleton / microtubule cytoskeleton organization / spindle / mitotic spindle / mitotic cell cycle / microtubule cytoskeleton / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / hydrolase activity / cell division / response to antibiotic / GTPase activity / GTP binding / ATP hydrolysis activity / ATP binding / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Moores, C.A. / von Loeffelholz, O. | |||||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: Cryo-EM Structure (4.5-Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance. Authors: Ottilie von Loeffelholz / Alejandro Peña / Douglas Robert Drummond / Robert Cross / Carolyn Ann Moores /  Abstract: Kinesin-5s are microtubule-dependent motors that drive spindle pole separation during mitosis. We used cryo-electron microscopy to determine the 4.5-Å resolution structure of the motor domain of the ...Kinesin-5s are microtubule-dependent motors that drive spindle pole separation during mitosis. We used cryo-electron microscopy to determine the 4.5-Å resolution structure of the motor domain of the fission yeast kinesin-5 Cut7 bound to fission yeast microtubules and explored the topology of the motor-microtubule interface and the susceptibility of the complex to drug binding. Despite their non-canonical architecture and mechanochemistry, Schizosaccharomyces pombe microtubules were stabilized by epothilone at the taxane binding pocket. The overall Cut7 footprint on the S. pombe microtubule surface is altered compared to mammalian tubulin microtubules because of their different polymer architectures. However, the core motor-microtubule interaction is tightly conserved, reflected in similar Cut7 ATPase activities on each microtubule type. AMPPNP-bound Cut7 adopts a kinesin-conserved ATP-like conformation including cover neck bundle formation. However, the Cut7 ATPase is not blocked by a mammalian-specific kinesin-5 inhibitor, consistent with the non-conserved sequence and structure of its loop5 insertion. #1: Journal: J Mol Biol / Year: 2020 Title: Corrigendum to "Cryo-EM Structure (4.5 Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance" [J. Mol. Biol. 431 (2019) 864-872] ...Title: Corrigendum to "Cryo-EM Structure (4.5 Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance" [J. Mol. Biol. 431 (2019) 864-872] https://doi.org/10.1016/j.jmb.2019.01.011. Authors: Ottilie von Loeffelholz / Alejandro Peña / Douglas Robert Drummond / Robert Cross / Carolyn Ann Moores /    | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6s8m.cif.gz 6s8m.cif.gz | 230.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6s8m.ent.gz pdb6s8m.ent.gz | 177.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6s8m.json.gz 6s8m.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s8/6s8m https://data.pdbj.org/pub/pdb/validation_reports/s8/6s8m ftp://data.pdbj.org/pub/pdb/validation_reports/s8/6s8m ftp://data.pdbj.org/pub/pdb/validation_reports/s8/6s8m | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3527MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | |
| Experimental dataset #1 | Data reference:  10.1016/j.jmb.2019.01.011 10.1016/j.jmb.2019.01.011 |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 3 types, 3 molecules KBA
| #1: Protein | Mass: 48219.750 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: cut7, SPAC25G10.07c / Production host:  |
|---|---|
| #2: Protein | Mass: 49518.359 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: nda3, alp12, SPBC26H8.07c / Production host:  |
| #3: Protein | Mass: 51203.180 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: nda2, SPBC16A3.15c / Production host:  |
-Non-polymers , 5 types, 5 molecules 


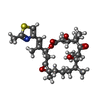





| #4: Chemical | ChemComp-MG / |
|---|---|
| #5: Chemical | ChemComp-ANP / |
| #6: Chemical | ChemComp-GDP / |
| #7: Chemical | ChemComp-EPB / |
| #8: Chemical | ChemComp-GTP / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Microtubule decorated with Cut7 motor domain and stabilized by epothilone-B Type: COMPLEX / Entity ID: #1-#3 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 6.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI POLARA 300 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Cs: 2.3 mm |
| Image recording | Electron dose: 30 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.12_2829: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Details: inside Frealign reconstruction procedure / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 33007 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj





















