+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ela | ||||||
|---|---|---|---|---|---|---|---|
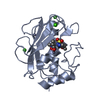
| Title | Crystal structure of MMP12 in complex with inhibitor BE4. | ||||||
 Components Components | Macrophage metalloelastase | ||||||
 Keywords Keywords | HYDROLASE / metzincin / carboxylate inhibitor alternative zinc-binding groups | ||||||
| Function / homology |  Function and homology information Function and homology informationmacrophage elastase / negative regulation of endothelial cell-matrix adhesion / bronchiole development / positive regulation of epithelial cell proliferation involved in wound healing / elastin catabolic process / regulation of defense response to virus by host / positive regulation of type I interferon-mediated signaling pathway / wound healing, spreading of epidermal cells / negative regulation of type I interferon-mediated signaling pathway / lung alveolus development ...macrophage elastase / negative regulation of endothelial cell-matrix adhesion / bronchiole development / positive regulation of epithelial cell proliferation involved in wound healing / elastin catabolic process / regulation of defense response to virus by host / positive regulation of type I interferon-mediated signaling pathway / wound healing, spreading of epidermal cells / negative regulation of type I interferon-mediated signaling pathway / lung alveolus development / Collagen degradation / response to amyloid-beta / collagen catabolic process / positive regulation of interferon-alpha production / extracellular matrix disassembly / core promoter sequence-specific DNA binding / collagen binding / Degradation of the extracellular matrix / extracellular matrix organization / extracellular matrix / metalloendopeptidase activity / cellular response to virus / protein import into nucleus / endopeptidase activity / sequence-specific DNA binding / serine-type endopeptidase activity / calcium ion binding / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / proteolysis / extracellular space / extracellular region / zinc ion binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.485 Å MOLECULAR REPLACEMENT / Resolution: 1.485 Å | ||||||
 Authors Authors | Ciccone, L. / Tepshi, L. / Nuti, E. / Rossello, A. / Stura, E.A. | ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2018 Journal: J. Med. Chem. / Year: 2018Title: Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies. Authors: Nuti, E. / Cuffaro, D. / Bernardini, E. / Camodeca, C. / Panelli, L. / Chaves, S. / Ciccone, L. / Tepshi, L. / Vera, L. / Orlandini, E. / Nencetti, S. / Stura, E.A. / Santos, M.A. / Dive, V. / Rossello, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ela.cif.gz 6ela.cif.gz | 316.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ela.ent.gz pdb6ela.ent.gz | 256 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ela.json.gz 6ela.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/el/6ela https://data.pdbj.org/pub/pdb/validation_reports/el/6ela ftp://data.pdbj.org/pub/pdb/validation_reports/el/6ela ftp://data.pdbj.org/pub/pdb/validation_reports/el/6ela | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6eknC  6enmC  6eoxC  6esmC  5i4oS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 17557.568 Da / Num. of mol.: 4 / Mutation: F171D, K241A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cell: macrophage / Gene: MMP12, HME / Production host: Homo sapiens (human) / Cell: macrophage / Gene: MMP12, HME / Production host:  |
|---|
-Non-polymers , 6 types, 893 molecules 

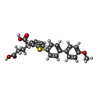








| #2: Chemical | ChemComp-ZN / #3: Chemical | ChemComp-CA / #4: Chemical | ChemComp-B9Z / ( #5: Chemical | ChemComp-DIO / #6: Chemical | ChemComp-EDO / | #7: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44.13 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: protein solution: hMMP12 at 314 micro-M + 10 milli-M acetohydroxamate + 1 milli-M BE4, 10% DMSO precipitant: 38% PEG 4K, 0.16 M imidazole piperidine,15% Dioxane, pH 8.5. Cryoprotectant: 40% ...Details: protein solution: hMMP12 at 314 micro-M + 10 milli-M acetohydroxamate + 1 milli-M BE4, 10% DMSO precipitant: 38% PEG 4K, 0.16 M imidazole piperidine,15% Dioxane, pH 8.5. Cryoprotectant: 40% CM15 (12.5 % diethylene glycol + 12.5 % ethylene glycol + 12.5 % MPD + 12.5 % glycerol + 12.5 % 1,2-propanediol + 12.5 % 1,4-dioxane + 12.5 mM NDSB 201), 25% PEG 6K, 100 milli-M TRIS HCl, pH 7.0 PH range: 7-8.5 / Temp details: cooled incubator |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: cryostream |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 2 / Wavelength: 0.976254 Å / Beamline: PROXIMA 2 / Wavelength: 0.976254 Å |
| Detector | Type: DECTRIS EIGER X 9M / Detector: PIXEL / Date: Jun 30, 2016 / Details: microfocus |
| Radiation | Monochromator: mirror / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.976254 Å / Relative weight: 1 |
| Reflection | Resolution: 1.48→43.44 Å / Num. obs: 100074 / % possible obs: 99.4 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 6.67 % / CC1/2: 0.997 / Rrim(I) all: 0.137 / Rsym value: 0.127 / Net I/σ(I): 7.85 |
| Reflection shell | Resolution: 1.48→1.57 Å / Redundancy: 6.51 % / Mean I/σ(I) obs: 1.68 / Num. unique obs: 15656 / CC1/2: 0.874 / Rrim(I) all: 0.85 / Rsym value: 0.78 / % possible all: 96.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5I4O Resolution: 1.485→38.041 Å / SU ML: 0.2 / Cross valid method: THROUGHOUT / σ(F): 1.01 / Phase error: 26.58 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.485→38.041 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj









