Entry Database : PDB / ID : 2ovxTitle MMP-9 active site mutant with barbiturate inhibitor Matrix metalloproteinase-9 (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2 Å Authors Tochowicz, A. / Bode, W. / Maskos, K. / Goettig, P. Journal : J.Mol.Biol. / Year : 2007Title : Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.Authors : Tochowicz, A. / Maskos, K. / Huber, R. / Oltenfreiter, R. / Dive, V. / Yiotakis, A. / Zanda, M. / Bode, W. / Goettig, P. History Deposition Feb 15, 2007 Deposition site / Processing site Revision 1.0 Jun 19, 2007 Provider / Type Revision 1.1 May 1, 2008 Group Revision 1.2 Jul 13, 2011 Group / Version format complianceRevision 1.3 Aug 16, 2017 Group / Source and taxonomy / Category / pdbx_unobs_or_zero_occ_atomsRevision 1.4 Oct 20, 2021 Group / Database references / Derived calculationsCategory database_2 / pdbx_struct_conn_angle ... database_2 / pdbx_struct_conn_angle / pdbx_unobs_or_zero_occ_atoms / struct_conn / struct_ref_seq_dif / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id Revision 1.5 Aug 30, 2023 Group / Refinement descriptionCategory / chem_comp_bond / pdbx_initial_refinement_model
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å
MOLECULAR REPLACEMENT / Resolution: 2 Å  Authors
Authors Citation
Citation Journal: J.Mol.Biol. / Year: 2007
Journal: J.Mol.Biol. / Year: 2007 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2ovx.cif.gz
2ovx.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2ovx.ent.gz
pdb2ovx.ent.gz PDB format
PDB format 2ovx.json.gz
2ovx.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ov/2ovx
https://data.pdbj.org/pub/pdb/validation_reports/ov/2ovx ftp://data.pdbj.org/pub/pdb/validation_reports/ov/2ovx
ftp://data.pdbj.org/pub/pdb/validation_reports/ov/2ovx


 Links
Links Assembly
Assembly
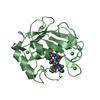

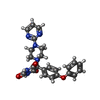
 Components
Components Homo sapiens (human) / Gene: MMP9, CLG4B / Production host:
Homo sapiens (human) / Gene: MMP9, CLG4B / Production host: 








 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site: MPG/DESY, HAMBURG
SYNCHROTRON / Site: MPG/DESY, HAMBURG  / Beamline: BW6 / Wavelength: 1.05
/ Beamline: BW6 / Wavelength: 1.05  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj














