[English] 日本語
 Yorodumi
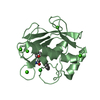
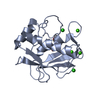
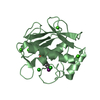
Yorodumi- PDB-3shi: Crystal structure of human MMP1 catalytic domain at 2.2 A resolution -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3shi | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human MMP1 catalytic domain at 2.2 A resolution | ||||||
 Components Components | Interstitial collagenase | ||||||
 Keywords Keywords | HYDROLASE / matrix metalloproteinase / paramagnetic restraints / paramagnetic tag / lanthanides / protein refinement / residual dipolar couplings | ||||||
| Function / homology |  Function and homology information Function and homology informationinterstitial collagenase / cellular response to UV-A / Basigin interactions / Activation of Matrix Metalloproteinases / Collagen degradation / collagen catabolic process / extracellular matrix disassembly / Degradation of the extracellular matrix / extracellular matrix organization / positive regulation of protein-containing complex assembly ...interstitial collagenase / cellular response to UV-A / Basigin interactions / Activation of Matrix Metalloproteinases / Collagen degradation / collagen catabolic process / extracellular matrix disassembly / Degradation of the extracellular matrix / extracellular matrix organization / positive regulation of protein-containing complex assembly / metalloendopeptidase activity / extracellular matrix / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / peptidase activity / Interleukin-4 and Interleukin-13 signaling / endopeptidase activity / serine-type endopeptidase activity / proteolysis / extracellular region / zinc ion binding Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Bertini, I. / Calderone, V. / Cerofolini, L. / Fragai, M. / Geraldes, C.F.G.C. / Hermann, P. / Luchinat, C. / Parigi, G. / Teixeira, J. | ||||||
 Citation Citation |  Journal: Febs Lett. / Year: 2012 Journal: Febs Lett. / Year: 2012Title: The catalytic domain of MMP-1 studied through tagged lanthanides. Authors: Bertini, I. / Calderone, V. / Cerofolini, L. / Fragai, M. / Geraldes, C.F. / Hermann, P. / Luchinat, C. / Parigi, G. / Teixeira, J.M. #1:  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself. Authors: Lovejoy, B. / Hassell, A.M. / Luther, M.A. / Weigl, D. / Jordan, S.R. #2:  Journal: Proteins / Year: 1994 Journal: Proteins / Year: 1994Title: 1.56 A structure of mature truncated human fibroblast collagenase. Authors: Spurlino, J.C. / Smallwood, A.M. / Carlton, D.D. / Banks, T.M. / Vavra, K.J. / Johnson, J.S. / Cook, E.R. / Falvo, J. / Wahl, R.C. / Pulvino, T.A. #3:  Journal: Nat.Struct.Biol. / Year: 1999 Journal: Nat.Struct.Biol. / Year: 1999Title: Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors. Authors: Lovejoy, B. / Welch, A.R. / Carr, S. / Luong, C. / Broka, C. / Hendricks, R.T. / Campbell, J.A. / Walker, K.A. / Martin, R. / Van Wart, H. / Browner, M.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3shi.cif.gz 3shi.cif.gz | 116.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3shi.ent.gz pdb3shi.ent.gz | 89 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3shi.json.gz 3shi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sh/3shi https://data.pdbj.org/pub/pdb/validation_reports/sh/3shi ftp://data.pdbj.org/pub/pdb/validation_reports/sh/3shi ftp://data.pdbj.org/pub/pdb/validation_reports/sh/3shi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  966cS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| 2 | 
| |||||||||
| 3 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 17457.984 Da / Num. of mol.: 3 / Fragment: UNP residues 106-261 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MMP1, CLG / Production host: Homo sapiens (human) / Gene: MMP1, CLG / Production host:  #2: Chemical | ChemComp-ZN / #3: Chemical | ChemComp-CA / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.13 Å3/Da / Density % sol: 60.67 % |
|---|---|
| Crystal grow | pH: 8.5 / Details: 30% PEG8000, 0.1 M Tris-HCl, pH 8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source: SEALED TUBE / Type: OXFORD DIFFRACTION ENHANCE ULTRA / Wavelength: 1.5406 |
| Detector | Type: OXFORD ONYX CCD / Detector: CCD / Date: Jul 26, 2005 / Details: mirrors |
| Radiation | Monochromator: graphite / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5406 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→81.65 Å / Num. all: 32923 / Num. obs: 32923 / % possible obs: 99.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3 % / Biso Wilson estimate: 40.1 Å2 / Rmerge(I) obs: 0.066 / Rsym value: 0.066 / Net I/σ(I): 9.1 |
| Reflection shell | Resolution: 2.2→2.32 Å / Redundancy: 3 % / Rmerge(I) obs: 0.49 / Mean I/σ(I) obs: 1.8 / Rsym value: 0.49 / % possible all: 99.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 966C Resolution: 2.2→81.65 Å / Cor.coef. Fo:Fc: 0.944 / Cor.coef. Fo:Fc free: 0.901 / Isotropic thermal model: ISOTROPIC, / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.258 / ESU R Free: 0.233 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT U VALUES : REFINED INDIVIDUALLY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.576 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→81.65 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.257 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj










