[English] 日本語
 Yorodumi
Yorodumi- EMDB-6777: Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6777 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | ||||||||||||
 Map data Map data | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cas13a / CRISPR / RNA BINDING PROTEIN-RNA COMPLEX | ||||||||||||
| Function / homology | : / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / RNA binding / CRISPR-associated endoribonuclease Cas13a Function and homology information Function and homology information | ||||||||||||
| Biological species |  Leptotrichia buccalis (bacteria) / Leptotrichia buccalis (bacteria) /  Leptotrichia buccalis (strain ATCC 14201 / DSM 1135 / JCM 12969 / NCTC 10249 / C-1013-b) (bacteria) / synthetic construct (others) Leptotrichia buccalis (strain ATCC 14201 / DSM 1135 / JCM 12969 / NCTC 10249 / C-1013-b) (bacteria) / synthetic construct (others) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Zhang X / Wang Y | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Authors: Liang Liu / Xueyan Li / Jun Ma / Zongqiang Li / Lilan You / Jiuyu Wang / Min Wang / Xinzheng Zhang / Yanli Wang /  Abstract: Cas13a, a type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, degrades invasive RNAs targeted by CRISPR RNA (crRNA) and has potential applications in RNA technology. To understand how Cas13a ...Cas13a, a type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, degrades invasive RNAs targeted by CRISPR RNA (crRNA) and has potential applications in RNA technology. To understand how Cas13a is activated to cleave RNA, we have determined the crystal structure of Leptotrichia buccalis (Lbu) Cas13a bound to crRNA and its target RNA, as well as the cryo-EM structure of the LbuCas13a-crRNA complex. The crRNA-target RNA duplex binds in a positively charged central channel of the nuclease (NUC) lobe, and Cas13a protein and crRNA undergo a significant conformational change upon target RNA binding. The guide-target RNA duplex formation triggers HEPN1 domain to move toward HEPN2 domain, activating the HEPN catalytic site of Cas13a protein, which subsequently cleaves both single-stranded target and collateral RNAs in a non-specific manner. These findings reveal how Cas13a of type VI CRISPR-Cas systems defend against RNA phages and set the stage for its development as a tool for RNA manipulation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6777.map.gz emd_6777.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6777-v30.xml emd-6777-v30.xml emd-6777.xml emd-6777.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6777.png emd_6777.png | 271.4 KB | ||
| Filedesc metadata |  emd-6777.cif.gz emd-6777.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6777 http://ftp.pdbj.org/pub/emdb/structures/EMD-6777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6777 | HTTPS FTP |
-Related structure data
| Related structure data |  5xwyMC  5xwpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6777.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6777.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
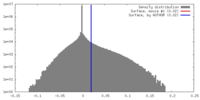
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : LbuCas13a-crRNA binary complex
| Entire | Name: LbuCas13a-crRNA binary complex |
|---|---|
| Components |
|
-Supramolecule #1: LbuCas13a-crRNA binary complex
| Supramolecule | Name: LbuCas13a-crRNA binary complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Leptotrichia buccalis (bacteria) / Strain: ATCC 14201 Leptotrichia buccalis (bacteria) / Strain: ATCC 14201 |
-Macromolecule #1: A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a
| Macromolecule | Name: A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leptotrichia buccalis (strain ATCC 14201 / DSM 1135 / JCM 12969 / NCTC 10249 / C-1013-b) (bacteria) Leptotrichia buccalis (strain ATCC 14201 / DSM 1135 / JCM 12969 / NCTC 10249 / C-1013-b) (bacteria)Strain: ATCC 14201 / DSM 1135 / JCM 12969 / NCTC 10249 / C-1013-b |
| Molecular weight | Theoretical: 138.555156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVTKVGGIS HKKYTSEGRL VKSESEENRT DERLSALLNM RLDMYIKNPS STETKENQKR IGKLKKFFSN KMVYLKDNTL SLKNGKKEN IDREYSETDI LESDVRDKKN FAVLKKIYLN ENVNSEELEV FRNDIKKKLN KINSLKYSFE KNKANYQKIN E NNIEKVEG ...String: MKVTKVGGIS HKKYTSEGRL VKSESEENRT DERLSALLNM RLDMYIKNPS STETKENQKR IGKLKKFFSN KMVYLKDNTL SLKNGKKEN IDREYSETDI LESDVRDKKN FAVLKKIYLN ENVNSEELEV FRNDIKKKLN KINSLKYSFE KNKANYQKIN E NNIEKVEG KSKRNIIYDY YRESAKRDAY VSNVKEAFDK LYKEEDIAKL VLEIENLTKL EKYKIREFYH EIIGRKNDKE NF AKIIYEE IQNVNNMKEL IEKVPDMSEL KKSQVFYKYY LDKEELNDKN IKYAFCHFVE IEMSQLLKNY VYKRLSNISN DKI KRIFEY QNLKKLIENK LLNKLDTYVR NCGKYNYYLQ DGEIATSDFI ARNRQNEAFL RNIIGVSSVA YFSLRNILET ENEN DITGR MRGKTVKNNK GEEKYVSGEV DKIYNENKKN EVKENLKMFY SYDFNMDNKN EIEDFFANID EAISSIRHGI VHFNL ELEG KDIFAFKNIA PSEISKKMFQ NEINEKKLKL KIFRQLNSAN VFRYLEKYKI LNYLKRTRFE FVNKNIPFVP SFTKLY SRI DDLKNSLGIY WKTPKTNDDN KTKEIIDAQI YLLKNIYYGE FLNYFMSNNG NFFEISKEII ELNKNDKRNL KTGFYKL QK FEDIQEKIPK EYLANIQSLY MINAGNQDEE EKDTYIDFIQ KIFLKGFMTY LANNGRLSLI YIGSDEETNT SLAEKKQE F DKFLKKYEQN NNIKIPYEIN EFLREIKLGN ILKYTERLNM FYLILKLLNH KELTNLKGSL EKYQSANKEE AFSDQLELI NLLNLDNNRV TEDFELEADE IGKFLDFNGN KVKDNKELKK FDTNKIYFDG ENIIKHRAFY NIKKYGMLNL LEKIADKAGY KISIEELKK YSNKKNEIEK NHKMQENLHR KYARPRKDEK FTDEDYESYK QAIENIEEYT HLKNKVEFNE LNLLQGLLLR I LHRLVGYT SIWERDLRFR LKGEFPENQY IEEIFNFENK KNVKYKGGQI VEKYIKFYKE LHQNDEVKIN KYSSANIKVL KQ EKKDLYI ANYIAAFNYI PHAEISLLEV LENLRKLLSY DRKLKNAVMK SVVDILKEYG FVATFKIGAD KKIGIQTLES EKI VHLKNL KKKKLMTDRN SEELCKLVKI MFEYKMEEKK SEN UniProtKB: CRISPR-associated endoribonuclease Cas13a |
-Macromolecule #2: RNA (59-MER)
| Macromolecule | Name: RNA (59-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 18.869348 KDa |
| Sequence | String: GGACCACCCC AAAAAUGAAG GGGACUAAAA CACAAAUCUA UCUGAAUAAA CUCUUCUUC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)