+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6687 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure for Hepatitis A virus empty particle | |||||||||
 Map data Map data | Cryo-EM map for HAV empty particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HAV / Neutralizing mechanism / Receptor recognition / Viral entry / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell mitochondrial outer membrane / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / host multivesicular body / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport ...host cell mitochondrial outer membrane / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / host multivesicular body / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / RNA helicase activity / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / symbiont entry into host cell / virion attachment to host cell / structural molecule activity / proteolysis / RNA binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |   Hepatitis A virus Hepatitis A virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Wang X / Zhu L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site. Authors: Xiangxi Wang / Ling Zhu / Minghao Dang / Zhongyu Hu / Qiang Gao / Shuai Yuan / Yao Sun / Bo Zhang / Jingshan Ren / Abhay Kotecha / Thomas S Walter / Junzhi Wang / Elizabeth E Fry / David I Stuart / Zihe Rao /   Abstract: Hepatitis A virus (HAV) infects ∼1.4 million people annually and, although there is a vaccine, there are no licensed therapeutic drugs. HAV is unusually stable (making disinfection problematic) and ...Hepatitis A virus (HAV) infects ∼1.4 million people annually and, although there is a vaccine, there are no licensed therapeutic drugs. HAV is unusually stable (making disinfection problematic) and little is known of how it enters cells and releases its RNA. Here we report a potent HAV-specific monoclonal antibody, R10, which neutralizes HAV infection by blocking attachment to the host cell. High-resolution cryo-EM structures of HAV full and empty particles and of the complex of HAV with R10 Fab reveal the atomic details of antibody binding and point to a receptor recognition site at the pentamer interface. These results, together with our observation that the R10 Fab destabilizes the capsid, suggest the use of a receptor mimic mechanism to neutralize virus infection, providing new opportunities for therapeutic intervention. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6687.map.gz emd_6687.map.gz | 167 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6687-v30.xml emd-6687-v30.xml emd-6687.xml emd-6687.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6687.png emd_6687.png | 237.7 KB | ||
| Filedesc metadata |  emd-6687.cif.gz emd-6687.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6687 http://ftp.pdbj.org/pub/emdb/structures/EMD-6687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6687 | HTTPS FTP |
-Validation report
| Summary document |  emd_6687_validation.pdf.gz emd_6687_validation.pdf.gz | 744.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6687_full_validation.pdf.gz emd_6687_full_validation.pdf.gz | 744.4 KB | Display | |
| Data in XML |  emd_6687_validation.xml.gz emd_6687_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_6687_validation.cif.gz emd_6687_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6687 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6687 | HTTPS FTP |
-Related structure data
| Related structure data |  5wtfMC  6686C  6688C  5wteC  5wtgC  5wthC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6687.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6687.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map for HAV empty particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
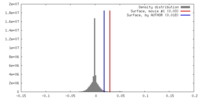
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hepatitis A virus
| Entire | Name:   Hepatitis A virus Hepatitis A virus |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis A virus
| Supramolecule | Name: Hepatitis A virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12092 / Sci species name: Hepatitis A virus / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 300.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis A virus / Organ: Homo sapiens Hepatitis A virus / Organ: Homo sapiens |
| Molecular weight | Theoretical: 25.16134 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: VGAITTIEDP VLAKKVPETF PELKPGESRH TSDHMSIYKF MGRSHFLCTF TFNSNNKEYT FPITLSSTSN PPHGLPSTLR WFFNLFQLY RGPLDLTIII TGATDVDGMA WFTPVGLAVD TPWVEKESAL QIDYKTALGA VRFNTRRTGN IQIRLPWYSY L YAVSGALD ...String: VGAITTIEDP VLAKKVPETF PELKPGESRH TSDHMSIYKF MGRSHFLCTF TFNSNNKEYT FPITLSSTSN PPHGLPSTLR WFFNLFQLY RGPLDLTIII TGATDVDGMA WFTPVGLAVD TPWVEKESAL QIDYKTALGA VRFNTRRTGN IQIRLPWYSY L YAVSGALD GLGDKTDSTF GLVSIQIANY NHSDEYLSFS CYLSVTEQSE FYFPRAPLNS NAMLST |
-Macromolecule #2: VP0
| Macromolecule | Name: VP0 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis A virus / Organ: Homo sapiens Hepatitis A virus / Organ: Homo sapiens |
| Molecular weight | Theoretical: 22.765881 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: ASYFTSVDQS SVHTAEVGSH QIEPLKTSVD KPGSKKTQGE KFFLIHSARW LTTHALFHEV AKLDVVKLLY NEQFAVQGLL RYHTYARFG IEIQVQINPT PFQQGGLICA MVPGDQSYGS IASLTVYPHG LLNCNINNVV RIKVPFIYTR GAYHFKDPQY P VWELTIRV ...String: ASYFTSVDQS SVHTAEVGSH QIEPLKTSVD KPGSKKTQGE KFFLIHSARW LTTHALFHEV AKLDVVKLLY NEQFAVQGLL RYHTYARFG IEIQVQINPT PFQQGGLICA MVPGDQSYGS IASLTVYPHG LLNCNINNVV RIKVPFIYTR GAYHFKDPQY P VWELTIRV WSELNIGTGT SAYTSLNVLA RFTDLELHGL TPLST |
-Macromolecule #3: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis A virus Hepatitis A virus |
| Molecular weight | Theoretical: 27.835693 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: MMRNETRVST TENVVNLSNY EDARAKMSFA LDQEDWKSDP SQGGGIKITH FTTWTSIPTL AAQFPFNASD SVGQQIKVIP VDPYFFQMT NTNPDQKCIT ALASICQMFC FWRGDLVFDF QVFPTKYHSG RLLFCFVPGN ELIDVTGITL KQATTAPCAV M DIAGVQST ...String: MMRNETRVST TENVVNLSNY EDARAKMSFA LDQEDWKSDP SQGGGIKITH FTTWTSIPTL AAQFPFNASD SVGQQIKVIP VDPYFFQMT NTNPDQKCIT ALASICQMFC FWRGDLVFDF QVFPTKYHSG RLLFCFVPGN ELIDVTGITL KQATTAPCAV M DIAGVQST LRFRVPWISD TPYRVNRYTK EAHQKGEYTA IGKLIVYCYN RLTSPSNVAH HVRVNVYLSA INLECFAPLY HA MDVTTQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS Buffer |
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK III / Details: blot for 3s seconds before plunging. |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-25 / Number grids imaged: 4 / Number real images: 500 / Average exposure time: 1.8 sec. / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 150 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-5wtf: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)