+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-6209 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of 20S supercomplex determined by single particle cryoelectron microscopy, state IIIb | |||||||||
 マップデータ マップデータ | Map of 20S supercomplex, state IIIb. This map is unsharpened and unfiltered. The map was normalized using the program MAPMAN. | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | vesicle trafficking | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報soluble NSF attachment protein activity / Intra-Golgi traffic / Retrograde transport at the Trans-Golgi-Network / COPI-dependent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / trans-Golgi Network Vesicle Budding / BLOC-1 complex / SNARE complex disassembly / regulation of delayed rectifier potassium channel activity / myosin head/neck binding ...soluble NSF attachment protein activity / Intra-Golgi traffic / Retrograde transport at the Trans-Golgi-Network / COPI-dependent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / trans-Golgi Network Vesicle Budding / BLOC-1 complex / SNARE complex disassembly / regulation of delayed rectifier potassium channel activity / myosin head/neck binding / exocytic insertion of neurotransmitter receptor to postsynaptic membrane / Other interleukin signaling / synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex / synaptobrevin 2-SNAP-25-syntaxin-1a complex / extrinsic component of presynaptic membrane / synaptic vesicle fusion to presynaptic active zone membrane / synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex / Lysosome Vesicle Biogenesis / positive regulation of norepinephrine secretion / Glutamate Neurotransmitter Release Cycle / Norepinephrine Neurotransmitter Release Cycle / COPII-mediated vesicle transport / positive regulation of catecholamine secretion / Acetylcholine Neurotransmitter Release Cycle / Serotonin Neurotransmitter Release Cycle / GABA synthesis, release, reuptake and degradation / Dopamine Neurotransmitter Release Cycle / Golgi Associated Vesicle Biogenesis / zymogen granule membrane / regulated exocytosis / ribbon synapse / presynaptic dense core vesicle exocytosis / synaptic vesicle docking / regulation of synaptic vesicle priming / storage vacuole / regulation of establishment of protein localization / response to gravity / protein-containing complex disassembly / vesicle-mediated transport in synapse / positive regulation of calcium ion-dependent exocytosis / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / calcium ion-regulated exocytosis of neurotransmitter / vesicle fusion / eosinophil degranulation / vesicle docking / chloride channel inhibitor activity / secretion by cell / ATP-dependent protein disaggregase activity / SNARE complex / SNAP receptor activity / Cargo recognition for clathrin-mediated endocytosis / regulation of exocytosis / regulation of vesicle-mediated transport / Clathrin-mediated endocytosis / LGI-ADAM interactions / calcium-ion regulated exocytosis / hormone secretion / intra-Golgi vesicle-mediated transport / actomyosin / positive regulation of intracellular protein transport / Golgi to plasma membrane protein transport / apical protein localization / Golgi stack / neurotransmitter secretion / positive regulation of hormone secretion / neurotransmitter receptor internalization / protein localization to membrane / ATP-dependent protein binding / neuron projection terminus / positive regulation of ATP-dependent activity / vesicle-fusing ATPase / regulation of synaptic vesicle recycling / insulin secretion / syntaxin binding / neurotransmitter transport / syntaxin-1 binding / clathrin-coated vesicle / SNARE complex assembly / positive regulation of neurotransmitter secretion / Neutrophil degranulation / endosomal transport / synaptic vesicle priming / regulation of synapse assembly / postsynaptic cytosol / myosin binding / positive regulation of receptor recycling / regulation of neuron projection development / positive regulation of exocytosis / modulation of excitatory postsynaptic potential / associative learning / exocytosis / synaptic vesicle exocytosis / protein sumoylation / positive regulation of excitatory postsynaptic potential / voltage-gated potassium channel activity / synaptic vesicle endocytosis / endomembrane system / calcium channel inhibitor activity / long-term memory / response to glucose 類似検索 - 分子機能 | |||||||||
| 生物種 |   | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 8.2 Å | |||||||||
 データ登録者 データ登録者 | Zhao M / Wu S / Zhou Q / Vivona S / Cipriano DJ / Cheng Y / Brunger AT | |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2015 ジャーナル: Nature / 年: 2015タイトル: Mechanistic insights into the recycling machine of the SNARE complex. 著者: Minglei Zhao / Shenping Wu / Qiangjun Zhou / Sandro Vivona / Daniel J Cipriano / Yifan Cheng / Axel T Brunger /  要旨: Evolutionarily conserved SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptors) proteins form a complex that drives membrane fusion in eukaryotes. The ATPase NSF (N- ...Evolutionarily conserved SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptors) proteins form a complex that drives membrane fusion in eukaryotes. The ATPase NSF (N-ethylmaleimide sensitive factor), together with SNAPs (soluble NSF attachment protein), disassembles the SNARE complex into its protein components, making individual SNAREs available for subsequent rounds of fusion. Here we report structures of ATP- and ADP-bound NSF, and the NSF/SNAP/SNARE (20S) supercomplex determined by single-particle electron cryomicroscopy at near-atomic to sub-nanometre resolution without imposing symmetry. Large, potentially force-generating, conformational differences exist between ATP- and ADP-bound NSF. The 20S supercomplex exhibits broken symmetry, transitioning from six-fold symmetry of the NSF ATPase domains to pseudo four-fold symmetry of the SNARE complex. SNAPs interact with the SNARE complex with an opposite structural twist, suggesting an unwinding mechanism. The interfaces between NSF, SNAPs, and SNAREs exhibit characteristic electrostatic patterns, suggesting how one NSF/SNAP species can act on many different SNARE complexes. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_6209.map.gz emd_6209.map.gz | 6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-6209-v30.xml emd-6209-v30.xml emd-6209.xml emd-6209.xml | 17.4 KB 17.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_6209.png emd_6209.png | 122.2 KB | ||
| その他 |  emd_6209_additional_1.map.gz emd_6209_additional_1.map.gz | 7.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6209 http://ftp.pdbj.org/pub/emdb/structures/EMD-6209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6209 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_6209_validation.pdf.gz emd_6209_validation.pdf.gz | 370 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_6209_full_validation.pdf.gz emd_6209_full_validation.pdf.gz | 369.6 KB | 表示 | |
| XML形式データ |  emd_6209_validation.xml.gz emd_6209_validation.xml.gz | 5.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6209 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6209 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6209 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6209 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  3j99MC  6204C  6205C  6206C  6207C  6208C  6210C  3j94C  3j95C  3j96C  3j97C  3j98C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_6209.map.gz / 形式: CCP4 / 大きさ: 7.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_6209.map.gz / 形式: CCP4 / 大きさ: 7.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Map of 20S supercomplex, state IIIb. This map is unsharpened and unfiltered. The map was normalized using the program MAPMAN. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.4312 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
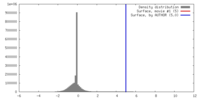
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-添付マップデータ: emd 6209 additional 1.map
| ファイル | emd_6209_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
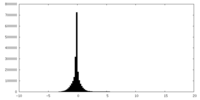
| 投影像・断面図 |
| ||||||||||||
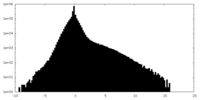
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : 20S supercomplex consisting of truncated neuronal SNARE complex, ...
| 全体 | 名称: 20S supercomplex consisting of truncated neuronal SNARE complex, alpha-SNAP, and N-ethylmaleimide sensitive factor (NSF) |
|---|---|
| 要素 |
|
-超分子 #1000: 20S supercomplex consisting of truncated neuronal SNARE complex, ...
| 超分子 | 名称: 20S supercomplex consisting of truncated neuronal SNARE complex, alpha-SNAP, and N-ethylmaleimide sensitive factor (NSF) タイプ: sample / ID: 1000 集合状態: One hexamer of NSF + four alpha-SNAP molecules + one SNARE complex Number unique components: 5 |
|---|---|
| 分子量 | 理論値: 660 KDa |
-分子 #1: N-ethylmaleimide sensitive factor
| 分子 | 名称: N-ethylmaleimide sensitive factor / タイプ: protein_or_peptide / ID: 1 / Name.synonym: NSF / コピー数: 6 / 集合状態: hexamer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  別称: Chinese hamster |
| 分子量 | 理論値: 83 KDa |
| 組換発現 | 生物種:  |
| 配列 | UniProtKB: Vesicle-fusing ATPase |
-分子 #2: alpha Soluble NSF Attachment Protein
| 分子 | 名称: alpha Soluble NSF Attachment Protein / タイプ: protein_or_peptide / ID: 2 / Name.synonym: alpha-SNAP / コピー数: 4 / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 33 KDa |
| 組換発現 | 生物種:  |
| 配列 | UniProtKB: Alpha-soluble NSF attachment protein |
-分子 #3: Syntaxin-1A
| 分子 | 名称: Syntaxin-1A / タイプ: protein_or_peptide / ID: 3 / Name.synonym: Stx-1A / コピー数: 1 / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 8 KDa |
| 組換発現 | 生物種:  |
| 配列 | UniProtKB: Syntaxin-1A |
-分子 #4: Synaptobrevin-2
| 分子 | 名称: Synaptobrevin-2 / タイプ: protein_or_peptide / ID: 4 / Name.synonym: Syb-2, VAMP-2 / コピー数: 1 / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 8 KDa |
| 組換発現 | 生物種:  |
| 配列 | UniProtKB: Vesicle-associated membrane protein 2 |
-分子 #5: Synaptosomal-associated protein 25
| 分子 | 名称: Synaptosomal-associated protein 25 / タイプ: protein_or_peptide / ID: 5 / Name.synonym: SNAP-25 / コピー数: 1 / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 16 KDa |
| 組換発現 | 生物種:  |
| 配列 | UniProtKB: Synaptosomal-associated protein 25 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 15 mg/mL |
|---|---|
| 緩衝液 | pH: 8 詳細: 50 mM Tris-Cl, 150 mM NaCl, 1 mM AMPPNP, 1 mM EDTA, 1 mM DTT, 0.05% v/v Nonident P-40 |
| グリッド | 詳細: Holey carbon on top of 400 mesh copper grid |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 90 K / 装置: FEI VITROBOT MARK I / 手法: Blot for 3.5 seconds before plunging. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI POLARA 300 |
|---|---|
| 日付 | 2014年1月28日 |
| 撮影 | カテゴリ: CCD / フィルム・検出器のモデル: GATAN K2 (4k x 4k) / 平均電子線量: 44 e/Å2 詳細: Gatan K2 Summit in super-resolution counting mode. Motion correction as described in Li et al. (2013) Nature Methods. |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.3 mm / 最大 デフォーカス(公称値): -2.8 µm / 最小 デフォーカス(公称値): -1.8 µm / 倍率(公称値): 31000 |
| 試料ステージ | 試料ホルダーモデル: OTHER |
| 実験機器 |  モデル: Tecnai Polara / 画像提供: FEI Company |
- 画像解析
画像解析
| 詳細 | 3D classification, refinement, and reconstruction were performed using RELION. |
|---|---|
| CTF補正 | 詳細: Each particle |
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 8.2 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: RELION / 使用した粒子像数: 14991 |
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - Chain ID: A |
|---|---|
| ソフトウェア | 名称: Chimera, PHENIX |
| 詳細 | D2 domain of NSF was from crystal structure 1NSF. D1 domain of NSF was from related entry EMD-6204. N domain of NSF was from crystal structure 1QCS. aSNAP was a homology model. SNARE complex was from crystal structure 1N7S. |
| 精密化 | 空間: RECIPROCAL / プロトコル: FLEXIBLE FIT / 当てはまり具合の基準: R-factor |
| 得られたモデル |  PDB-3j99: |
-原子モデル構築 2
| 初期モデル | PDB ID: Chain - Chain ID: A |
|---|---|
| ソフトウェア | 名称: Chimera, PHENIX |
| 詳細 | D2 domain of NSF was from crystal structure 1NSF. D1 domain of NSF was from related entry EMD-6204. N domain of NSF was from crystal structure 1QCS. aSNAP was a homology model. SNARE complex was from crystal structure 1N7S. |
| 精密化 | 空間: RECIPROCAL / プロトコル: FLEXIBLE FIT / 当てはまり具合の基準: R-factor |
| 得られたモデル |  PDB-3j99: |
-原子モデル構築 3
| 初期モデル | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D |
|---|---|
| ソフトウェア | 名称: Chimera, PHENIX |
| 詳細 | D2 domain of NSF was from crystal structure 1NSF. D1 domain of NSF was from related entry EMD-6204. N domain of NSF was from crystal structure 1QCS. aSNAP was a homology model. SNARE complex was from crystal structure 1N7S. |
| 精密化 | 空間: RECIPROCAL / プロトコル: FLEXIBLE FIT / 当てはまり具合の基準: R-factor |
| 得られたモデル |  PDB-3j99: |
 ムービー
ムービー コントローラー
コントローラー



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)