[English] 日本語
 Yorodumi
Yorodumi- EMDB-5859: Structures of Cas9 endonucleases reveal RNA-mediated conformation... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5859 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation | |||||||||
 Map data Map data | Reconstruction of Cas9 loaded with crRNA:tracrRNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / crRNA / tracrRNA / Cas9 / genome engineering / bacterial immunity | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / 3'-5' exonuclease activity / DNA endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Jinek M / Jiang F / Taylor DW / Sternberg SH / Kaya E / Ma E / Anders C / Hauer M / Zhou K / Lin S ...Jinek M / Jiang F / Taylor DW / Sternberg SH / Kaya E / Ma E / Anders C / Hauer M / Zhou K / Lin S / Kaplan M / Iavarone AT / Charpentier E / Nogales E / Doudna JA | |||||||||
 Citation Citation |  Journal: Science / Year: 2014 Journal: Science / Year: 2014Title: Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Authors: Martin Jinek / Fuguo Jiang / David W Taylor / Samuel H Sternberg / Emine Kaya / Enbo Ma / Carolin Anders / Michael Hauer / Kaihong Zhou / Steven Lin / Matias Kaplan / Anthony T Iavarone / ...Authors: Martin Jinek / Fuguo Jiang / David W Taylor / Samuel H Sternberg / Emine Kaya / Enbo Ma / Carolin Anders / Michael Hauer / Kaihong Zhou / Steven Lin / Matias Kaplan / Anthony T Iavarone / Emmanuelle Charpentier / Eva Nogales / Jennifer A Doudna /  Abstract: Type II CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems use an RNA-guided DNA endonuclease, Cas9, to generate double-strand breaks in invasive DNA ...Type II CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems use an RNA-guided DNA endonuclease, Cas9, to generate double-strand breaks in invasive DNA during an adaptive bacterial immune response. Cas9 has been harnessed as a powerful tool for genome editing and gene regulation in many eukaryotic organisms. We report 2.6 and 2.2 angstrom resolution crystal structures of two major Cas9 enzyme subtypes, revealing the structural core shared by all Cas9 family members. The architectures of Cas9 enzymes define nucleic acid binding clefts, and single-particle electron microscopy reconstructions show that the two structural lobes harboring these clefts undergo guide RNA-induced reorientation to form a central channel where DNA substrates are bound. The observation that extensive structural rearrangements occur before target DNA duplex binding implicates guide RNA loading as a key step in Cas9 activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5859.map.gz emd_5859.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5859-v30.xml emd-5859-v30.xml emd-5859.xml emd-5859.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5859.png emd_5859.png | 240.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5859 http://ftp.pdbj.org/pub/emdb/structures/EMD-5859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5859 | HTTPS FTP |
-Validation report
| Summary document |  emd_5859_validation.pdf.gz emd_5859_validation.pdf.gz | 79 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5859_full_validation.pdf.gz emd_5859_full_validation.pdf.gz | 78.1 KB | Display | |
| Data in XML |  emd_5859_validation.xml.gz emd_5859_validation.xml.gz | 492 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5859 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5859 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5859 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5859 | HTTPS FTP |
-Related structure data
| Related structure data |  5858C  5860C  4cmpC  4cmqC  4ogcC  4ogeC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5859.map.gz / Format: CCP4 / Size: 5.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5859.map.gz / Format: CCP4 / Size: 5.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Cas9 loaded with crRNA:tracrRNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
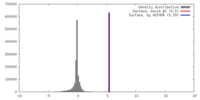
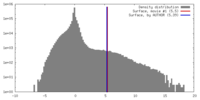
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cas9 loaded with crRNA:tracrRNA
| Entire | Name: Cas9 loaded with crRNA:tracrRNA |
|---|---|
| Components |
|
-Supramolecule #1000: Cas9 loaded with crRNA:tracrRNA
| Supramolecule | Name: Cas9 loaded with crRNA:tracrRNA / type: sample / ID: 1000 Oligomeric state: one Cas9 binds to one crRNA:tracRNA heteroduplex Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 196 KDa |
-Macromolecule #1: CRISPR-associated endonuclease Cas9
| Macromolecule | Name: CRISPR-associated endonuclease Cas9 / type: protein_or_peptide / ID: 1 / Name.synonym: Cas9 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 159 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: CRISPR-associated endonuclease Cas9/Csn1 GO: defense response to virus, maintenance of CRISPR repeat elements, 3'-5' exonuclease activity, DNA endonuclease activity, RNA binding, DNA binding InterPro: INTERPRO: IPR010145, INTERPRO: IPR025978, HNH nuclease |
-Macromolecule #2: CRISPR RNA
| Macromolecule | Name: CRISPR RNA / type: rna / ID: 2 / Name.synonym: crRNA Details: Forms complete guide RNA by hybridization with tracrRNA. Classification: OTHER / Structure: OTHER / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 14 KDa |
| Sequence | String: GUGAUAAGUG GAAUGCCAUG GUUUUAGAGC UAUGCUGUUU UG |
-Macromolecule #3: trans-activating CRISPR RNA
| Macromolecule | Name: trans-activating CRISPR RNA / type: rna / ID: 3 / Name.synonym: tracrRNA Details: Forms complete guide RNA by hybridization with crRNA. Made using T7 RNA polymerase. Classification: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 24 KDa |
| Sequence | String: GGACAGCAUA GCAAGUUAAA AUAAGGCUAG UCCGUUAUCA ACUUGAAAAA GUGGCACCGA GUCGGUGCUU UUU |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Tris-Cl pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol |
| Staining | Type: NEGATIVE Details: After adsorption for 1 min, we stained the samples consecutively with six droplets of 2% (w/v) uranyl acetate solution, gently blotted off the residual stain, and air-dried the sample in a fume hood. |
| Grid | Details: glow-discharged 400 mesh continuous carbon grids |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Temperature | Average: 78 K |
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism was corrected at 210,000 times magnification Legacy - Electron beam tilt params: 0 |
| Details | Data acquired using Leginon. |
| Date | May 10, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 297 / Average electron dose: 20 e/Å2 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: -1.4 µm / Nominal defocus min: -0.5 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder: Room temperature single tilt / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Details | Particles were selected using template based picking in Appion. |
|---|---|
| CTF correction | Details: whole micrograph |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: OTHER / Software - Name: EMAN2, SPARX / Details: Image pre-processing performed in Appion. / Number images used: 36982 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)