[English] 日本語
 Yorodumi
Yorodumi- PDB-4bt0: MuB is an AAAplus ATPase that forms helical filaments to control ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4bt0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | ||||||
 Components Components | (TRANSCRIPTIONAL REGULATOR) x 2 | ||||||
 Keywords Keywords | TRANSCRIPTION / AAA+ ATPASE / DNA TRANSPOSITION / NUCLEOPROTEIN FILAMENT / SYMMETRY MISMATCH | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA-binding transcription activator activity / phosphorelay signal transduction system / cis-regulatory region sequence-specific DNA binding / protein-DNA complex / positive regulation of DNA-templated transcription / ATP hydrolysis activity / ATP binding / metal ion binding / identical protein binding Similarity search - Function | ||||||
| Biological species |  ENTEROBACTERIA PHAGE MU (virus) ENTEROBACTERIA PHAGE MU (virus) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 17 Å | ||||||
 Authors Authors | Mizuno, N. / Dramicanin, M. / Mizuuchi, M. / Adam, J. / Wang, Y. / Han, Y.W. / Yang, W. / Steven, A.C. / Mizuuchi, K. / Ramon-Maiques, S. | ||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2013 Journal: Proc Natl Acad Sci U S A / Year: 2013Title: MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. Authors: Naoko Mizuno / Marija Dramićanin / Michiyo Mizuuchi / Julia Adam / Yi Wang / Yong-Woon Han / Wei Yang / Alasdair C Steven / Kiyoshi Mizuuchi / Santiago Ramón-Maiques /  Abstract: MuB is an ATP-dependent nonspecific DNA-binding protein that regulates the activity of the MuA transposase and captures target DNA for transposition. Mechanistic understanding of MuB function has ...MuB is an ATP-dependent nonspecific DNA-binding protein that regulates the activity of the MuA transposase and captures target DNA for transposition. Mechanistic understanding of MuB function has previously been hindered by MuB's poor solubility. Here we combine bioinformatic, mutagenic, biochemical, and electron microscopic analyses to unmask the structure and function of MuB. We demonstrate that MuB is an ATPase associated with diverse cellular activities (AAA+ ATPase) and forms ATP-dependent filaments with or without DNA. We also identify critical residues for MuB's ATPase, DNA binding, protein polymerization, and MuA interaction activities. Using single-particle electron microscopy, we show that MuB assembles into a helical filament, which binds the DNA in the axial channel. The helical parameters of the MuB filament do not match those of the coated DNA. Despite this protein-DNA symmetry mismatch, MuB does not deform the DNA duplex. These findings, together with the influence of MuB filament size on strand-transfer efficiency, lead to a model in which MuB-imposed symmetry transiently deforms the DNA at the boundary of the MuB filament and results in a bent DNA favored by MuA for transposition. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4bt0.cif.gz 4bt0.cif.gz | 63.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4bt0.ent.gz pdb4bt0.ent.gz | 45.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4bt0.json.gz 4bt0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4bt0_validation.pdf.gz 4bt0_validation.pdf.gz | 915.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4bt0_full_validation.pdf.gz 4bt0_full_validation.pdf.gz | 936.5 KB | Display | |
| Data in XML |  4bt0_validation.xml.gz 4bt0_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  4bt0_validation.cif.gz 4bt0_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bt/4bt0 https://data.pdbj.org/pub/pdb/validation_reports/bt/4bt0 ftp://data.pdbj.org/pub/pdb/validation_reports/bt/4bt0 ftp://data.pdbj.org/pub/pdb/validation_reports/bt/4bt0 | HTTPS FTP |
-Related structure data
| Related structure data |  2398MC 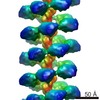 2395C 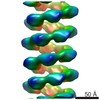 2400C  4bs1C  4bt1C C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 7
| ||||||||||||||||||||||||||||||||
| 2 |
| ||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||
| Symmetry | Helical symmetry: (Circular symmetry: 1 / Dyad axis: no / N subunits divisor: 1 / Num. of operations: 7 / Rise per n subunits: 9.01 Å / Rotation per n subunits: 66.2 °) | ||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 8562.894 Da / Num. of mol.: 1 / Fragment: AAAPLUS DOMAIN, RESIDUES 312-384 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  ENTEROBACTERIA PHAGE MU (virus) / Production host: ENTEROBACTERIA PHAGE MU (virus) / Production host:  |
|---|---|
| #2: Protein | Mass: 19397.400 Da / Num. of mol.: 1 / Fragment: AAAPLUS DOMAIN, RESIDUES 137-309 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  ENTEROBACTERIA PHAGE MU (virus) / Production host: ENTEROBACTERIA PHAGE MU (virus) / Production host:  |
| #3: Chemical |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: MUB FILAMENT WITH DNA / Type: COMPLEX |
|---|---|
| Buffer solution | Name: 30 MM TRISHCL PH 8.0, 0.3 M KCL, 5MM MGCL2, 1MM DTT, 1 MM ATP OR ATP-GAMMA-S pH: 8 Details: 30 MM TRISHCL PH 8.0, 0.3 M KCL, 5MM MGCL2, 1MM DTT, 1 MM ATP OR ATP-GAMMA-S |
| Specimen | Conc.: 0.07 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: HOLEY CARBON |
| Vitrification | Instrument: FEI VITROBOT MARK II / Cryogen name: ETHANE / Details: LIQUID ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: FEI/PHILIPS CM200FEG / Date: Apr 27, 2008 |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 120 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 120 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 38000 X / Nominal defocus max: 3000 nm / Nominal defocus min: 1500 nm |
| Specimen holder | Temperature: 82 K |
| Image recording | Electron dose: 15 e/Å2 / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) |
| Image scans | Num. digital images: 109 |
- Processing
Processing
| EM software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: PHASE-FLIPPING | |||||||||||||||
| 3D reconstruction | Method: IHRSR / Resolution: 17 Å / Nominal pixel size: 2.8 Å / Actual pixel size: 2.8 Å Magnification calibration: SUBMISSION BASED ON EXPERIMENTAL DATA FROM EMDB EMD-2398. (DEPOSITION ID: 11698) Symmetry type: HELICAL | |||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL Details: METHOD--RIGID BODY REFINEMENT PROTOCOL--ELECTRON MICROSCPY | |||||||||||||||
| Atomic model building | PDB-ID: 1NY6 Accession code: 1NY6 / Source name: PDB / Type: experimental model | |||||||||||||||
| Refinement | Highest resolution: 17 Å | |||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 17 Å
|
 Movie
Movie Controller
Controller








 PDBj
PDBj




