[English] 日本語
 Yorodumi
Yorodumi- EMDB-2395: MuB is an AAA+ ATPase that forms helical filaments to control tar... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2395 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition | |||||||||
 Map data Map data | Reconstruction of MuB filament without DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / DNA transposition / Mu phage / nucleoprotein filament / symmetry mismatch | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-binding transcription activator activity / phosphorelay signal transduction system / cis-regulatory region sequence-specific DNA binding / protein-DNA complex / positive regulation of DNA-templated transcription / ATP binding / metal ion binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Enterobacteria phage Mu (virus) Enterobacteria phage Mu (virus) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Mizuno N / Dramicanin M / Mizuuchi M / Adam J / Wang Y / Han YW / Yang W / Steven AC / Mizuuchi K / Ramon-Maiques S | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2013 Journal: Proc Natl Acad Sci U S A / Year: 2013Title: MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. Authors: Naoko Mizuno / Marija Dramićanin / Michiyo Mizuuchi / Julia Adam / Yi Wang / Yong-Woon Han / Wei Yang / Alasdair C Steven / Kiyoshi Mizuuchi / Santiago Ramón-Maiques /  Abstract: MuB is an ATP-dependent nonspecific DNA-binding protein that regulates the activity of the MuA transposase and captures target DNA for transposition. Mechanistic understanding of MuB function has ...MuB is an ATP-dependent nonspecific DNA-binding protein that regulates the activity of the MuA transposase and captures target DNA for transposition. Mechanistic understanding of MuB function has previously been hindered by MuB's poor solubility. Here we combine bioinformatic, mutagenic, biochemical, and electron microscopic analyses to unmask the structure and function of MuB. We demonstrate that MuB is an ATPase associated with diverse cellular activities (AAA+ ATPase) and forms ATP-dependent filaments with or without DNA. We also identify critical residues for MuB's ATPase, DNA binding, protein polymerization, and MuA interaction activities. Using single-particle electron microscopy, we show that MuB assembles into a helical filament, which binds the DNA in the axial channel. The helical parameters of the MuB filament do not match those of the coated DNA. Despite this protein-DNA symmetry mismatch, MuB does not deform the DNA duplex. These findings, together with the influence of MuB filament size on strand-transfer efficiency, lead to a model in which MuB-imposed symmetry transiently deforms the DNA at the boundary of the MuB filament and results in a bent DNA favored by MuA for transposition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2395.map.gz emd_2395.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2395-v30.xml emd-2395-v30.xml emd-2395.xml emd-2395.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2395.png emd_2395.png | 146.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2395 http://ftp.pdbj.org/pub/emdb/structures/EMD-2395 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2395 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2395 | HTTPS FTP |
-Validation report
| Summary document |  emd_2395_validation.pdf.gz emd_2395_validation.pdf.gz | 238.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2395_full_validation.pdf.gz emd_2395_full_validation.pdf.gz | 237.3 KB | Display | |
| Data in XML |  emd_2395_validation.xml.gz emd_2395_validation.xml.gz | 4.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2395 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2395 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2395 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2395 | HTTPS FTP |
-Related structure data
| Related structure data |  4bs1MC  2398C 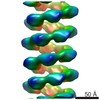 2400C  4bt0C  4bt1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2395.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2395.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of MuB filament without DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : MuB filament without DNA
| Entire | Name: MuB filament without DNA |
|---|---|
| Components |
|
-Supramolecule #1000: MuB filament without DNA
| Supramolecule | Name: MuB filament without DNA / type: sample / ID: 1000 / Oligomeric state: helical filament / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 35 KDa / Theoretical: 35 KDa |
-Macromolecule #1: MuB AAA+ ATPase
| Macromolecule | Name: MuB AAA+ ATPase / type: protein_or_peptide / ID: 1 / Oligomeric state: helical assembly / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage Mu (virus) Enterobacteria phage Mu (virus) |
| Molecular weight | Experimental: 35 KDa / Theoretical: 35 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.07 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 30 mM TrisHCl pH 8.0, 0.15 M KCl, 5mM MgCl2, 1mM DTT, 1 mM ATP or ATP-gamma-S |
| Grid | Details: 300 mesh quantifoil R2/2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 90 K / Instrument: FEI VITROBOT MARK II / Method: Blot for 5 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Min: 80 K / Max: 85 K / Average: 82 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Date | Apr 21, 2008 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Number real images: 129 / Average electron dose: 15 e/Å2 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: side entry / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | IHRSR |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 9.01 Å Applied symmetry - Helical parameters - Δ&Phi: 66.2 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: spider, bsoft, eman |
| CTF correction | Details: phase-flipping |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















