+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-4348 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of a late human pre-40S ribosomal subunit - State B | |||||||||||||||
 マップデータ マップデータ | ||||||||||||||||
 試料 試料 |
| |||||||||||||||
 キーワード キーワード | 40S / pre-40S / ribosome biogenesis / RIBOSOME | |||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報regulation of protein localization to nucleolus / trophectodermal cell differentiation / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / positive regulation of respiratory burst involved in inflammatory response / nucleolus organization / negative regulation of RNA splicing / U3 snoRNA binding / neural crest cell differentiation / positive regulation of ubiquitin-protein transferase activity / negative regulation of bicellular tight junction assembly ...regulation of protein localization to nucleolus / trophectodermal cell differentiation / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / positive regulation of respiratory burst involved in inflammatory response / nucleolus organization / negative regulation of RNA splicing / U3 snoRNA binding / neural crest cell differentiation / positive regulation of ubiquitin-protein transferase activity / negative regulation of bicellular tight junction assembly / snoRNA binding / rRNA modification in the nucleus and cytosol / preribosome, small subunit precursor / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / negative regulation of ubiquitin protein ligase activity / laminin receptor activity / Ribosomal scanning and start codon recognition / Translation initiation complex formation / fibroblast growth factor binding / monocyte chemotaxis / TOR signaling / Protein hydroxylation / SARS-CoV-1 modulates host translation machinery / cellular response to ethanol / mTORC1-mediated signalling / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Eukaryotic Translation Termination / negative regulation of ubiquitin-dependent protein catabolic process / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / ubiquitin ligase inhibitor activity / Viral mRNA Translation / negative regulation of respiratory burst involved in inflammatory response / positive regulation of signal transduction by p53 class mediator / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / regulation of translational fidelity / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Protein methylation / Nuclear events stimulated by ALK signaling in cancer / positive regulation of cell cycle / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rough endoplasmic reticulum / laminin binding / translation regulator activity / ribosomal small subunit export from nucleus / translation initiation factor binding / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / MDM2/MDM4 family protein binding / visual perception / RNA endonuclease activity / antiviral innate immune response / liver regeneration / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / stress granule assembly / cytosolic ribosome / Resolution of Sister Chromatid Cohesion / stem cell proliferation / cellular response to leukemia inhibitory factor / positive regulation of translation / erythrocyte differentiation / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / innate immune response in mucosa / mRNA 3'-UTR binding / maturation of SSU-rRNA / neural tube closure / translational initiation / small-subunit processome / RHO GTPases Activate Formins / maintenance of translational fidelity / response to insulin / mRNA 5'-UTR binding / modification-dependent protein catabolic process / GABA-ergic synapse / response to virus / Regulation of expression of SLITs and ROBOs / protein tag activity / RMTs methylate histone arginines / apical part of cell / cytoplasmic ribonucleoprotein granule / rRNA processing / osteoblast differentiation / Separation of Sister Chromatids / antimicrobial humoral immune response mediated by antimicrobial peptide / glucose homeostasis / antibacterial humoral response / presynapse / ribosome biogenesis / chromosome / ribosome binding / cell body 類似検索 - 分子機能 | |||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.0 Å | |||||||||||||||
 データ登録者 データ登録者 | Ameismeier M / Cheng J | |||||||||||||||
| 資金援助 |  ドイツ, 4件 ドイツ, 4件
| |||||||||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2018 ジャーナル: Nature / 年: 2018タイトル: Visualizing late states of human 40S ribosomal subunit maturation. 著者: Michael Ameismeier / Jingdong Cheng / Otto Berninghausen / Roland Beckmann /  要旨: The formation of eukaryotic ribosomal subunits extends from the nucleolus to the cytoplasm and entails hundreds of assembly factors. Despite differences in the pathways of ribosome formation, high- ...The formation of eukaryotic ribosomal subunits extends from the nucleolus to the cytoplasm and entails hundreds of assembly factors. Despite differences in the pathways of ribosome formation, high-resolution structural information has been available only from fungi. Here we present cryo-electron microscopy structures of late-stage human 40S assembly intermediates, representing one state reconstituted in vitro and five native states that range from nuclear to late cytoplasmic. The earliest particles reveal the position of the biogenesis factor RRP12 and distinct immature rRNA conformations that accompany the formation of the 40S subunit head. Molecular models of the late-acting assembly factors TSR1, RIOK1, RIOK2, ENP1, LTV1, PNO1 and NOB1 provide mechanistic details that underlie their contribution to a sequential 40S subunit assembly. The NOB1 architecture displays an inactive nuclease conformation that requires rearrangement of the PNO1-bound 3' rRNA, thereby coordinating the final rRNA folding steps with site 3 cleavage. | |||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_4348.map.gz emd_4348.map.gz | 17.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-4348-v30.xml emd-4348-v30.xml emd-4348.xml emd-4348.xml | 47.8 KB 47.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_4348_fsc.xml emd_4348_fsc.xml | 12.9 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_4348.png emd_4348.png | 159.9 KB | ||
| Filedesc metadata |  emd-4348.cif.gz emd-4348.cif.gz | 11 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4348 http://ftp.pdbj.org/pub/emdb/structures/EMD-4348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4348 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  6g4sMC  4337C  4349C  4350C  4351C  4352C  4353C  6g18C  6g4wC  6g51C  6g53C  6g5hC  6g5iC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_4348.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_4348.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
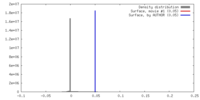
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Cryo-EM structure of a late human pre-40S ribosomal subunit - State B
+超分子 #1: Cryo-EM structure of a late human pre-40S ribosomal subunit - State B
+分子 #1: pre-18S ribosomal RNA
+分子 #2: 40S ribosomal protein S17
+分子 #3: 40S ribosomal protein SA
+分子 #4: 40S ribosomal protein S27
+分子 #5: 40S ribosomal protein S3a
+分子 #6: 40S ribosomal protein S2
+分子 #7: 40S ribosomal protein S28
+分子 #8: 40S ribosomal protein S4, X isoform
+分子 #9: 40S ribosomal protein S30
+分子 #10: 40S ribosomal protein S5
+分子 #11: 40S ribosomal protein S7
+分子 #12: 40S ribosomal protein S6
+分子 #13: 40S ribosomal protein S25
+分子 #14: 40S ribosomal protein S24
+分子 #15: RNA-binding protein NOB1
+分子 #16: RNA-binding protein PNO1
+分子 #17: 40S ribosomal protein S23
+分子 #18: Bystin
+分子 #19: 40S ribosomal protein S15a
+分子 #20: 40S ribosomal protein S21
+分子 #21: Pre-rRNA-processing protein TSR1 homolog
+分子 #22: Protein LTV1 homolog
+分子 #23: 40S ribosomal protein S19
+分子 #24: 40S ribosomal protein S18
+分子 #25: 40S ribosomal protein S16
+分子 #26: 40S ribosomal protein S15
+分子 #27: 40S ribosomal protein S14
+分子 #28: 40S ribosomal protein S13
+分子 #29: 40S ribosomal protein S11
+分子 #30: 40S ribosomal protein S9
+分子 #31: 40S ribosomal protein S8
+分子 #32: RRP12
+分子 #33: Unknown
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON II (4k x 4k) 平均電子線量: 2.5 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)