+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3crf | ||||||
|---|---|---|---|---|---|---|---|
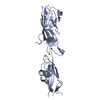
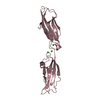
| Title | Electron Microscopy model of the Saf Pilus- Type B | ||||||
 Components Components | (Outer membrane protein) x 3 | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / SafA protein polymer / Type B Saf Pilus / Fibril Protein | ||||||
| Function / homology | Saf-pilin pilus formation protein / Saf-pilin pilus formation protein / Dr-adhesin superfamily / Adhesion domain superfamily / Prokaryotic membrane lipoprotein lipid attachment site profile. / Outer membrane protein Function and homology information Function and homology information | ||||||
| Biological species |  Salmonella typhimurium (bacteria) Salmonella typhimurium (bacteria) | ||||||
| Method | ELECTRON MICROSCOPY / helical reconstruction / negative staining / Resolution: 17 Å | ||||||
 Authors Authors | Salih, O. / Remaut, H. / Waksman, G. / Orlova, E.V. | ||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2008 Journal: J Mol Biol / Year: 2008Title: Structural analysis of the Saf pilus by electron microscopy and image processing. Authors: Osman Salih / Han Remaut / Gabriel Waksman / Elena V Orlova /  Abstract: Bacterial pili are important virulence factors involved in host cell attachment and/or biofilm formation, key steps in establishing and maintaining successful infection. Here we studied Salmonella ...Bacterial pili are important virulence factors involved in host cell attachment and/or biofilm formation, key steps in establishing and maintaining successful infection. Here we studied Salmonella atypical fimbriae (or Saf pili), formed by the conserved chaperone/usher pathway. In contrast to the well-established quaternary structure of typical/FGS-chaperone assembled, rod-shaped, chaperone/usher pili, little is known about the supramolecular organisation in atypical/FGL-chaperone assembled fimbriae. In our study, we have used negative stain electron microscopy and single-particle image analysis to determine the three-dimensional structure of the Salmonella typhimurium Saf pilus. Our results show atypical/FGL-chaperone assembled fimbriae are composed of highly flexible linear multi-subunit fibres that are formed by globular subunits connected to each other by short links giving a "beads on a string"-like appearance. Quantitative fitting of the atomic structure of the SafA pilus subunit into the electron density maps, in combination with linker modelling and energy minimisation, has enabled analysis of subunit arrangement and intersubunit interactions in the Saf pilus. Short intersubunit linker regions provide the molecular basis for flexibility of the Saf pilus by acting as molecular hinges allowing a large range of movement between consecutive subunits in the fibre. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3crf.cif.gz 3crf.cif.gz | 57.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3crf.ent.gz pdb3crf.ent.gz | 40.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3crf.json.gz 3crf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cr/3crf https://data.pdbj.org/pub/pdb/validation_reports/cr/3crf ftp://data.pdbj.org/pub/pdb/validation_reports/cr/3crf ftp://data.pdbj.org/pub/pdb/validation_reports/cr/3crf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1495MC  1494C  3creC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 12992.533 Da / Num. of mol.: 1 Fragment: core pilin domain, N-terminal deleted, UNP residues 48-170 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Salmonella typhimurium (bacteria) / Strain: LT2 / Gene: safA / Plasmid: PTRC99A / Production host: Salmonella typhimurium (bacteria) / Strain: LT2 / Gene: safA / Plasmid: PTRC99A / Production host:  |
|---|---|
| #2: Protein | Mass: 15270.004 Da / Num. of mol.: 1 / Fragment: core pilin domain, UNP residues 27-170 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Salmonella typhimurium (bacteria) / Strain: LT2 / Gene: safA / Plasmid: PTRC99A / Production host: Salmonella typhimurium (bacteria) / Strain: LT2 / Gene: safA / Plasmid: PTRC99A / Production host:  |
| #3: Protein/peptide | Mass: 2070.237 Da / Num. of mol.: 1 / Fragment: N-TERMINAL EXTENSION, UNP residues 27-45 / Source method: obtained synthetically / Details: SafANte-N-terminal extension / References: UniProt: Q8ZRK4 |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: helical reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Wild-type SafA pili / Type: COMPLEX Details: Stained with 2% ammonium molybdate. Type B Saf pilus. Fibre. |
|---|---|
| Buffer solution | Name: 10 mM TRIS-HCl (pH 8) and 10 mM NaCl / pH: 8 / Details: 10 mM TRIS-HCl (pH 8) and 10 mM NaCl |
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: YES / Vitrification applied: NO |
| EM staining | Type: NEGATIVE / Material: Ammonium Molybdate |
| Specimen support | Details: Continuous-carbon film |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: FEI TECNAI 10 / Date: Apr 23, 2004 |
|---|---|
| Electron gun | Electron source: TUNGSTEN HAIRPIN / Accelerating voltage: 100 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 44000 X / Nominal defocus max: 900 nm / Nominal defocus min: 400 nm / Cs: 2.4 mm |
| Image recording | Electron dose: 25 e/Å2 / Film or detector model: KODAK SO-163 FILM |
- Processing
Processing
| EM software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||
| 3D reconstruction | Method: Cross-common lines / Resolution: 17 Å / Num. of particles: 8500 / Actual pixel size: 1.591 Å / Symmetry type: POINT | ||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: RECIPROCAL / Target criteria: Correlation coefficient / Details: REFINEMENT PROTOCOL--Rigid body | ||||||||||||
| Atomic model building | PDB-ID: 2CO4 Accession code: 2CO4 / Source name: PDB / Type: experimental model | ||||||||||||
| Refinement step | Cycle: LAST
|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera













 PDBj
PDBj




