+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3874 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomogram of Ebola virus | ||||||||||||
 Map data Map data | A 4x binned representative tomogram of intact Ebola virus virions | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
| Method | electron tomography / cryo EM | ||||||||||||
 Authors Authors | Wan W / Kolesnikova L / Clarke M / Koehler A / Noda T / Becker S / Briggs JAG | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structure and assembly of the Ebola virus nucleocapsid. Authors: William Wan / Larissa Kolesnikova / Mairi Clarke / Alexander Koehler / Takeshi Noda / Stephan Becker / John A G Briggs /    Abstract: Ebola and Marburg viruses are filoviruses: filamentous, enveloped viruses that cause haemorrhagic fever. Filoviruses are within the order Mononegavirales, which also includes rabies virus, measles ...Ebola and Marburg viruses are filoviruses: filamentous, enveloped viruses that cause haemorrhagic fever. Filoviruses are within the order Mononegavirales, which also includes rabies virus, measles virus, and respiratory syncytial virus. Mononegaviruses have non-segmented, single-stranded negative-sense RNA genomes that are encapsidated by nucleoprotein and other viral proteins to form a helical nucleocapsid. The nucleocapsid acts as a scaffold for virus assembly and as a template for genome transcription and replication. Insights into nucleoprotein-nucleoprotein interactions have been derived from structural studies of oligomerized, RNA-encapsidating nucleoprotein, and cryo-electron microscopy of nucleocapsid or nucleocapsid-like structures. There have been no high-resolution reconstructions of complete mononegavirus nucleocapsids. Here we apply cryo-electron tomography and subtomogram averaging to determine the structure of Ebola virus nucleocapsid within intact viruses and recombinant nucleocapsid-like assemblies. These structures reveal the identity and arrangement of the nucleocapsid components, and suggest that the formation of an extended α-helix from the disordered carboxy-terminal region of nucleoprotein-core links nucleoprotein oligomerization, nucleocapsid condensation, RNA encapsidation, and accessory protein recruitment. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3874.map.gz emd_3874.map.gz | 80 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3874-v30.xml emd-3874-v30.xml emd-3874.xml emd-3874.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3874.png emd_3874.png | 224.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3874 http://ftp.pdbj.org/pub/emdb/structures/EMD-3874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3874 | HTTPS FTP |
-Validation report
| Summary document |  emd_3874_validation.pdf.gz emd_3874_validation.pdf.gz | 200.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3874_full_validation.pdf.gz emd_3874_full_validation.pdf.gz | 199.3 KB | Display | |
| Data in XML |  emd_3874_validation.xml.gz emd_3874_validation.xml.gz | 3.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3874 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3874 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3874.map.gz / Format: CCP4 / Size: 409.8 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_3874.map.gz / Format: CCP4 / Size: 409.8 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A 4x binned representative tomogram of intact Ebola virus virions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
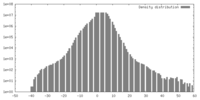
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ebola virus - Mayinga, Zaire, 1976
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Ebola virus - Mayinga, Zaire, 1976
| Supramolecule | Name: Ebola virus - Mayinga, Zaire, 1976 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Virus was isolated from infected VeroE6 cells. Purified viruses were fixed with paraformaldehyde. NCBI-ID: 128952 / Sci species name: Ebola virus - Mayinga, Zaire, 1976 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: Nucleocapsid / Diameter: 280.0 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: Virus was purified into Dulbecco's modified Eagle's medium (DMEM) with 4% paraformaldehyde |
|---|---|
| Grid | Model: C-flat 2/1 3C / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK II |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: UMC Utrecht / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: -10 eV / Energy filter - Upper energy threshold: 10 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3708 pixel / Digitization - Dimensions - Height: 3708 pixel / Digitization - Frames/image: 1-5 / Average exposure time: 2.0 sec. / Average electron dose: 2.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Frames were aligned using K2Align software, based off the MotionCorr algorithm. Tomograms were reconstructed with IMOD, using stripwise CTF-correction and weighted back projection. | ||||||
|---|---|---|---|---|---|---|---|
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 41 IMOD / Number images used: 41 | ||||||
| CTF correction | Software:
Details: CTF amplitude correction was performed during the wedge-weighted subtomogram averaging step. |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)