[English] 日本語
 Yorodumi
Yorodumi- EMDB-36451: The Cryo-EM structure of a heptameric CED-4/CED-3 catalytic complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Cryo-EM structure of a heptameric CED-4/CED-3 catalytic complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CED-4 / CED-3 catalytic domain / APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationBH1 domain binding / positive regulation of apoptotic process involved in development / regulation of development, heterochronic / caspase complex / positive regulation of synapse pruning / peptidase activator activity involved in apoptotic process / positive regulation of protein processing / caspase binding / embryonic morphogenesis / apoptotic process involved in development ...BH1 domain binding / positive regulation of apoptotic process involved in development / regulation of development, heterochronic / caspase complex / positive regulation of synapse pruning / peptidase activator activity involved in apoptotic process / positive regulation of protein processing / caspase binding / embryonic morphogenesis / apoptotic process involved in development / negative regulation of execution phase of apoptosis / actin filament depolymerization / activation of cysteine-type endopeptidase activity / embryo development ending in birth or egg hatching / regulation of cell size / muscle cell cellular homeostasis / BH3 domain binding / cysteine-type endopeptidase activator activity involved in apoptotic process / endopeptidase activator activity / regulation of cell adhesion / regulation of protein stability / ADP binding / activation of cysteine-type endopeptidase activity involved in apoptotic process / defense response to Gram-negative bacterium / positive regulation of apoptotic process / apoptotic process / negative regulation of apoptotic process / perinuclear region of cytoplasm / magnesium ion binding / protein-containing complex / mitochondrion / ATP binding / identical protein binding / membrane / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Li Y / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2023 Journal: Life Sci Alliance / Year: 2023Title: Structural insights into CED-3 activation. Authors: Yini Li / Lu Tian / Ying Zhang / Yigong Shi /  Abstract: In , onset of programmed cell death is marked with the activation of CED-3, a process that requires assembly of the CED-4 apoptosome. Activated CED-3 forms a holoenzyme with the CED-4 apoptosome to ...In , onset of programmed cell death is marked with the activation of CED-3, a process that requires assembly of the CED-4 apoptosome. Activated CED-3 forms a holoenzyme with the CED-4 apoptosome to cleave a wide range of substrates, leading to irreversible cell death. Despite decades of investigations, the underlying mechanism of CED-4-facilitated CED-3 activation remains elusive. Here, we report cryo-EM structures of the CED-4 apoptosome and three distinct CED-4/CED-3 complexes that mimic different activation stages for CED-3. In addition to the previously reported octamer in crystal structures, CED-4, alone or in complex with CED-3, exists in multiple oligomeric states. Supported by biochemical analyses, we show that the conserved CARD-CARD interaction promotes CED-3 activation, and initiation of programmed cell death is regulated by the dynamic organization of the CED-4 apoptosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36451.map.gz emd_36451.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36451-v30.xml emd-36451-v30.xml emd-36451.xml emd-36451.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36451.png emd_36451.png | 51.3 KB | ||
| Filedesc metadata |  emd-36451.cif.gz emd-36451.cif.gz | 5.5 KB | ||
| Others |  emd_36451_additional_1.map.gz emd_36451_additional_1.map.gz emd_36451_half_map_1.map.gz emd_36451_half_map_1.map.gz emd_36451_half_map_2.map.gz emd_36451_half_map_2.map.gz | 49.3 MB 49.6 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36451 http://ftp.pdbj.org/pub/emdb/structures/EMD-36451 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36451 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36451 | HTTPS FTP |
-Validation report
| Summary document |  emd_36451_validation.pdf.gz emd_36451_validation.pdf.gz | 728.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36451_full_validation.pdf.gz emd_36451_full_validation.pdf.gz | 728.1 KB | Display | |
| Data in XML |  emd_36451_validation.xml.gz emd_36451_validation.xml.gz | 12 KB | Display | |
| Data in CIF |  emd_36451_validation.cif.gz emd_36451_validation.cif.gz | 14.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36451 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36451 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36451 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36451 | HTTPS FTP |
-Related structure data
| Related structure data |  8jo0MC  8jnsC  8jolC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36451.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36451.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||
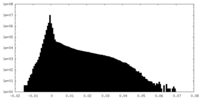
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_36451_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
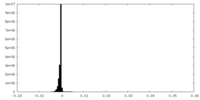
| Density Histograms |
-Half map: #2
| File | emd_36451_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
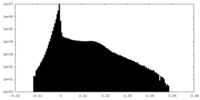
| Density Histograms |
-Half map: #1
| File | emd_36451_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
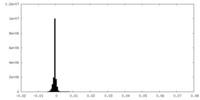
| Density Histograms |
- Sample components
Sample components
-Entire : Heptameric CED-4/CED-3 catalytic complex
| Entire | Name: Heptameric CED-4/CED-3 catalytic complex |
|---|---|
| Components |
|
-Supramolecule #1: Heptameric CED-4/CED-3 catalytic complex
| Supramolecule | Name: Heptameric CED-4/CED-3 catalytic complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cell death protein 4
| Macromolecule | Name: Cell death protein 4 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.933644 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLCEIECRAL STAHTRLIHD FEPRDALTYL EGKNIFTEDH SELISKMSTR LERIANFLRI YRRQASELGP LIDFFNYNNQ SHLADFLED YIDFAINEPD LLRPVVIAPQ F UniProtKB: Cell death protein 4 |
-Macromolecule #2: Cell death protein 4
| Macromolecule | Name: Cell death protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.953266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLCEIECRAL STAHTRLIHD FEPRDALTYL EGKNIFTEDH SELISKMSTR LERIANFLRI YRRQASELGP LIDFFNYNNQ SHLADFLED YIDFAINEPD LLRPVVIAPQ FSRQMLDRKL LLGNVPKQMT CYIREYHVDR VIKKLDEMCD LDSFFLFLHG R AGSGKSVI ...String: MLCEIECRAL STAHTRLIHD FEPRDALTYL EGKNIFTEDH SELISKMSTR LERIANFLRI YRRQASELGP LIDFFNYNNQ SHLADFLED YIDFAINEPD LLRPVVIAPQ FSRQMLDRKL LLGNVPKQMT CYIREYHVDR VIKKLDEMCD LDSFFLFLHG R AGSGKSVI ASQALSKSDQ LIGINYDSIV WLKDSGTAPK STFDLFTDIL LMLKSEDDLL NFPSVEHVTS VVLKRMICNA LI DRPNTLF VFDDVVQEET IRWAQELRLR CLVTTRDVEI SNAASQTCEF IEVTSLEIDE CYDFLEAYGM PMPVGEKEED VLN KTIELS SGNPATLMMF FKSCEPKTFE KMAQLNNKLE SRGLVGVECI TPYSYKSLAM ALQRCVEVLS DEDRSALAFA VVMP PGVDI PVKLWSCVIP VDICSNEEEQ LDDEVADRLK RLSKRGALLS GKRMPVLTFK IDHIIHMFLK HVVDAQTIAN GISIL EQRL LEIGNNNVSV PERHIPSHFQ KFRRSSASEM YPKTTEETVI RPEDFPKFMQ LHQKFYDSLK NFACC UniProtKB: Cell death protein 4 |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 7 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 7 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 67312 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X