+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3533 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of the 70S chloroplast ribosome | |||||||||
 Map data Map data | Cryo-EM reconstruction of the 70S chloroplast ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | chloroplast / translation / ribosome / cryo-EM | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast rRNA processing / plastid small ribosomal subunit / plastid translation / negative regulation of translational elongation / chloroplast envelope / mitochondrial large ribosomal subunit / mitochondrial small ribosomal subunit / mitochondrial translation / chloroplast stroma / plastid ...chloroplast rRNA processing / plastid small ribosomal subunit / plastid translation / negative regulation of translational elongation / chloroplast envelope / mitochondrial large ribosomal subunit / mitochondrial small ribosomal subunit / mitochondrial translation / chloroplast stroma / plastid / chloroplast thylakoid membrane / ribosomal small subunit binding / chloroplast / DNA-templated transcription termination / large ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / mRNA binding / mitochondrion / RNA binding Similarity search - Function | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
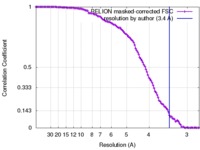
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Bieri P / Leibundgut M | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2017 Journal: EMBO J / Year: 2017Title: The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. Authors: Philipp Bieri / Marc Leibundgut / Martin Saurer / Daniel Boehringer / Nenad Ban /  Abstract: Chloroplasts are cellular organelles of plants and algae that are responsible for energy conversion and carbon fixation by the photosynthetic reaction. As a consequence of their endosymbiotic origin, ...Chloroplasts are cellular organelles of plants and algae that are responsible for energy conversion and carbon fixation by the photosynthetic reaction. As a consequence of their endosymbiotic origin, they still contain their own genome and the machinery for protein biosynthesis. Here, we present the atomic structure of the chloroplast 70S ribosome prepared from spinach leaves and resolved by cryo-EM at 3.4 Å resolution. The complete structure reveals the features of the 4.5S rRNA, which probably evolved by the fragmentation of the 23S rRNA, and all five plastid-specific ribosomal proteins. These proteins, required for proper assembly and function of the chloroplast translation machinery, bind and stabilize rRNA including regions that only exist in the chloroplast ribosome. Furthermore, the structure reveals plastid-specific extensions of ribosomal proteins that extensively remodel the mRNA entry and exit site on the small subunit as well as the polypeptide tunnel exit and the putative binding site of the signal recognition particle on the large subunit. The translation factor pY, involved in light- and temperature-dependent control of protein synthesis, is bound to the mRNA channel of the small subunit and interacts with 16S rRNA nucleotides at the A-site and P-site, where it protects the decoding centre and inhibits translation by preventing tRNA binding. The small subunit is locked by pY in a non-rotated state, in which the intersubunit bridges to the large subunit are stabilized. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3533.map.gz emd_3533.map.gz | 13.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3533-v30.xml emd-3533-v30.xml emd-3533.xml emd-3533.xml | 80.2 KB 80.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3533_fsc.xml emd_3533_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_3533.png emd_3533.png | 231.2 KB | ||
| Filedesc metadata |  emd-3533.cif.gz emd-3533.cif.gz | 16.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3533 http://ftp.pdbj.org/pub/emdb/structures/EMD-3533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3533 | HTTPS FTP |
-Related structure data
| Related structure data |  5mmmMC  3531C  3532C  5mmiC  5mmjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3533.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3533.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the 70S chloroplast ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Chloroplast 70S ribosome
+Supramolecule #1: Chloroplast 70S ribosome
+Macromolecule #1: 50S ribosomal protein L31
+Macromolecule #2: 50S ribosomal protein L32, chloroplastic
+Macromolecule #3: 50S ribosomal protein L33, chloroplastic
+Macromolecule #4: 50S ribosomal protein L34, chloroplastic
+Macromolecule #5: 50S ribosomal protein L35, chloroplastic
+Macromolecule #6: 50S ribosomal protein L36, chloroplastic
+Macromolecule #7: plastid ribosomal protein cL37, PSRP5
+Macromolecule #8: 50S ribosomal protein 6, chloroplastic
+Macromolecule #11: 50S ribosomal protein L2, chloroplastic
+Macromolecule #12: plastid ribosomal protein uL3c
+Macromolecule #13: plastid ribosomal protein uL4c
+Macromolecule #14: plastid ribosomal protein uL5c
+Macromolecule #15: plastid ribosomal protein uL6c
+Macromolecule #16: plastid ribosomal protein bL9c
+Macromolecule #17: plastid ribosomal protein uL10c
+Macromolecule #18: 50S ribosomal protein L11, chloroplastic
+Macromolecule #19: 50S ribosomal protein L13, chloroplastic
+Macromolecule #20: 50S ribosomal protein L14, chloroplastic
+Macromolecule #21: plastid ribosomal protein uL15c
+Macromolecule #22: 50S ribosomal protein L16, chloroplastic
+Macromolecule #23: plastid ribosomal protein uL14c
+Macromolecule #24: plastid ribosomal protein uL18c
+Macromolecule #25: 50S ribosomal protein L19, chloroplastic
+Macromolecule #26: 50S ribosomal protein L20, chloroplastic
+Macromolecule #27: 50S ribosomal protein L21, chloroplastic
+Macromolecule #28: 50S ribosomal protein L22, chloroplastic
+Macromolecule #29: 50S ribosomal protein L23, chloroplastic
+Macromolecule #30: plastid ribosomal protein uL24c
+Macromolecule #32: plastid ribosomal protein bL27c
+Macromolecule #33: plastid ribosomal protein bL28c
+Macromolecule #34: plastid ribosomal protein uL29c
+Macromolecule #36: plastid ribosomal protein bS1c
+Macromolecule #38: 30S ribosomal protein S2, chloroplastic
+Macromolecule #39: 30S ribosomal protein S3, chloroplastic
+Macromolecule #40: 30S ribosomal protein S4, chloroplastic
+Macromolecule #41: 30S ribosomal protein S5, chloroplastic
+Macromolecule #42: plastid ribosomal protein bS6c
+Macromolecule #43: 30S ribosomal protein S7, chloroplastic
+Macromolecule #44: 30S ribosomal protein S8, chloroplastic
+Macromolecule #45: plastid ribosomal protein uS9c
+Macromolecule #46: plastid ribosomal protein uS10c
+Macromolecule #47: 30S ribosomal protein S11, chloroplastic
+Macromolecule #48: 30S ribosomal protein S12, chloroplastic
+Macromolecule #49: plastid ribosomal protein uS13c
+Macromolecule #50: 30S ribosomal protein S14, chloroplastic
+Macromolecule #51: 30S ribosomal protein S15, chloroplastic
+Macromolecule #52: 30S ribosomal protein S16, chloroplastic
+Macromolecule #53: plastid ribosomal protein uS17c
+Macromolecule #54: 30S ribosomal protein S18, chloroplastic
+Macromolecule #55: 30S ribosomal protein S19 alpha, chloroplastic
+Macromolecule #56: plastid ribosomal protein bS20c
+Macromolecule #57: plastid ribosomal protein bS21c
+Macromolecule #58: 30S ribosomal protein 2, chloroplastic
+Macromolecule #59: 30S ribosomal protein 3, chloroplastic
+Macromolecule #60: 30S ribosomal protein S31, chloroplastic
+Macromolecule #61: Ribosome-binding factor PSRP1, chloroplastic
+Macromolecule #9: 23S ribosomal RNA
+Macromolecule #10: 5S ribosomal RNA
+Macromolecule #31: 4.5S ribosomal RNA
+Macromolecule #35: tRNA-Phe
+Macromolecule #37: 16S ribosomal RNA
+Macromolecule #62: ZINC ION
+Macromolecule #63: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Frames/image: 2-8 / Number real images: 2796 / Average exposure time: 0.77 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 90.8 |
|---|---|
| Output model |  PDB-5mmm: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)