+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3448 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA Polymerase I elongation complex 2 | |||||||||
 Map data Map data | RNA Polymerase I elongation complex 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA Polymerase / transcription | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase I preinitiation complex assembly / RNA Polymerase I Transcription Initiation / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape ...RNA polymerase I preinitiation complex assembly / RNA Polymerase I Transcription Initiation / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / termination of RNA polymerase III transcription / RNA Polymerase II Pre-transcription Events / RNA-templated transcription / Formation of TC-NER Pre-Incision Complex / regulation of cell size / RNA Polymerase I Promoter Escape / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / Gap-filling DNA repair synthesis and ligation in TC-NER / nucleolar large rRNA transcription by RNA polymerase I / transcription initiation at RNA polymerase I promoter / Estrogen-dependent gene expression / transcription by RNA polymerase III / Dual incision in TC-NER / RNA polymerase I complex / transcription elongation by RNA polymerase I / RNA polymerase III complex / tRNA transcription by RNA polymerase III / RNA polymerase II, core complex / transcription by RNA polymerase I / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / promoter-specific chromatin binding / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / peroxisome / ribosome biogenesis / nucleic acid binding / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / protein dimerization activity / nucleolus / negative regulation of transcription by RNA polymerase II / mitochondrion / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
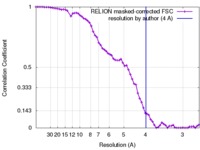
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Tafur L / Sadian Y | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2016 Journal: Mol Cell / Year: 2016Title: Molecular Structures of Transcribing RNA Polymerase I. Authors: Lucas Tafur / Yashar Sadian / Niklas A Hoffmann / Arjen J Jakobi / Rene Wetzel / Wim J H Hagen / Carsten Sachse / Christoph W Müller /  Abstract: RNA polymerase I (Pol I) is a 14-subunit enzyme that solely synthesizes pre-ribosomal RNA. Recently, the crystal structure of apo Pol I gave unprecedented insight into its molecular architecture. ...RNA polymerase I (Pol I) is a 14-subunit enzyme that solely synthesizes pre-ribosomal RNA. Recently, the crystal structure of apo Pol I gave unprecedented insight into its molecular architecture. Here, we present three cryo-EM structures of elongating Pol I, two at 4.0 Å and one at 4.6 Å resolution, and a Pol I open complex at 3.8 Å resolution. Two modules in Pol I mediate the narrowing of the DNA-binding cleft by closing the clamp domain. The DNA is bound by the clamp head and by the protrusion domain, allowing visualization of the upstream and downstream DNA duplexes in one of the elongation complexes. During formation of the Pol I elongation complex, the bridge helix progressively folds, while the A12.2 C-terminal domain is displaced from the active site. Our results reveal the conformational changes associated with elongation complex formation and provide additional insight into the Pol I transcription cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3448.map.gz emd_3448.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3448-v30.xml emd-3448-v30.xml emd-3448.xml emd-3448.xml | 37.8 KB 37.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3448_fsc.xml emd_3448_fsc.xml | 6.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_3448.png emd_3448.png | 101.1 KB | ||
| Filedesc metadata |  emd-3448.cif.gz emd-3448.cif.gz | 10.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3448 http://ftp.pdbj.org/pub/emdb/structures/EMD-3448 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3448 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3448 | HTTPS FTP |
-Related structure data
| Related structure data |  5m5yMC  3446C  3447C  3449C  5m5wC  5m5xC  5m64C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3448.map.gz / Format: CCP4 / Size: 20.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3448.map.gz / Format: CCP4 / Size: 20.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNA Polymerase I elongation complex 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : RNA Polymerase I elongation complex 2
+Supramolecule #1: RNA Polymerase I elongation complex 2
+Supramolecule #2: Transcription scaffold
+Macromolecule #1: DNA-directed RNA polymerase I subunit RPA190
+Macromolecule #2: DNA-directed RNA polymerase I subunit RPA135
+Macromolecule #3: DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #4: DNA-directed RNA polymerase I subunit RPA14
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerase I subunit RPA43
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase I subunit RPA12
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #11: DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #13: DNA-directed RNA polymerase I subunit RPA49
+Macromolecule #14: DNA-directed RNA polymerase I subunit RPA34
+Macromolecule #15: RNA
+Macromolecule #16: Non-template DNA
+Macromolecule #17: Template DNA
+Macromolecule #18: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK II Details: 2.5 ul of sample 15 seconds wait time Blot for 8 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-5m5y: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)