[English] 日本語
 Yorodumi
Yorodumi- EMDB-31950: Cryo-EM structure of Vaccinia virus scaffolding protein D13 with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31950 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Vaccinia virus scaffolding protein D13 with N-terminal polyhistidine tag | ||||||||||||
 Map data Map data | Cryo-EM single particle reconstruction on Vaccinia virus scaffold protein D13 trimer with N-terminal poly-histidine tag. The map has been sharpened and density-normalized. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Scaffold / capsid / double-jelly-roll / VIRAL PROTEIN | ||||||||||||
| Function / homology | Poxvirus rifampicin-resistance / Poxvirus rifampicin resistance protein / response to antibiotic / identical protein binding / membrane / Scaffold protein OPG125 Function and homology information Function and homology information | ||||||||||||
| Biological species |  Vaccinia virus WR / Vaccinia virus WR /  Vaccinia virus (strain Western Reserve) Vaccinia virus (strain Western Reserve) | ||||||||||||
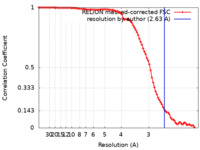
| Method | single particle reconstruction / cryo EM / Resolution: 2.63 Å | ||||||||||||
 Authors Authors | Wolf M / Hyun J | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Assembly mechanism of the pleomorphic immature poxvirus scaffold. Authors: Jaekyung Hyun / Hideyuki Matsunami / Tae Gyun Kim / Matthias Wolf /    Abstract: In Vaccinia virus (VACV), the prototype poxvirus, scaffold protein D13 forms a honeycomb-like lattice on the viral membrane that results in formation of the pleomorphic immature virion (IV). The ...In Vaccinia virus (VACV), the prototype poxvirus, scaffold protein D13 forms a honeycomb-like lattice on the viral membrane that results in formation of the pleomorphic immature virion (IV). The structure of D13 is similar to those of major capsid proteins that readily form icosahedral capsids in nucleocytoplasmic large DNA viruses (NCLDVs). However, the detailed assembly mechanism of the nonicosahedral poxvirus scaffold has never been understood. Here we show the cryo-EM structures of the D13 trimer and scaffold intermediates produced in vitro. The structures reveal that the displacement of the short N-terminal α-helix is critical for initiation of D13 self-assembly. The continuous curvature of the IV is mediated by electrostatic interactions that induce torsion between trimers. The assembly mechanism explains the semiordered capsid-like arrangement of D13 that is distinct from icosahedral NCLDVs. Our structures explain how a single protein can self-assemble into different capsid morphologies and represent a local exception to the universal Caspar-Klug theory of quasi-equivalence. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31950.map.gz emd_31950.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31950-v30.xml emd-31950-v30.xml emd-31950.xml emd-31950.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31950_fsc.xml emd_31950_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_31950.png emd_31950.png | 113.1 KB | ||
| Masks |  emd_31950_msk_1.map emd_31950_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31950.cif.gz emd-31950.cif.gz | 6.8 KB | ||
| Others |  emd_31950_additional_1.map.gz emd_31950_additional_1.map.gz emd_31950_additional_2.map.gz emd_31950_additional_2.map.gz emd_31950_half_map_1.map.gz emd_31950_half_map_1.map.gz emd_31950_half_map_2.map.gz emd_31950_half_map_2.map.gz | 49.6 MB 5.7 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31950 http://ftp.pdbj.org/pub/emdb/structures/EMD-31950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31950 | HTTPS FTP |
-Validation report
| Summary document |  emd_31950_validation.pdf.gz emd_31950_validation.pdf.gz | 726.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31950_full_validation.pdf.gz emd_31950_full_validation.pdf.gz | 725.6 KB | Display | |
| Data in XML |  emd_31950_validation.xml.gz emd_31950_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_31950_validation.cif.gz emd_31950_validation.cif.gz | 21.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31950 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31950 | HTTPS FTP |
-Related structure data
| Related structure data |  7vfeMC  7vfdC  7vffC  7vfgC  7vfhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31950.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31950.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM single particle reconstruction on Vaccinia virus scaffold protein D13 trimer with N-terminal poly-histidine tag. The map has been sharpened and density-normalized. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.036 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_31950_msk_1.map emd_31950_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Cryo-EM single particle reconstruction on Vaccinia virus scaffold...
| File | emd_31950_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM single particle reconstruction on Vaccinia virus scaffold protein D13 trimer with N-terminal poly-histidine tag. The map is unsharpened and density-normalized. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Cryo-EM single particle reconstruction on Vaccinia virus scaffold...
| File | emd_31950_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM single particle reconstruction on Vaccinia virus scaffold protein D13 trimer with N-terminal poly-histidine tag. The map is density-modified and density-normalized. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM single particle reconstruction on Vaccinia virus scaffold...
| File | emd_31950_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM single particle reconstruction on Vaccinia virus scaffold protein D13 trimer with N-terminal poly-histidine tag (half map 1). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM single particle reconstruction on Vaccinia virus scaffold...
| File | emd_31950_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM single particle reconstruction on Vaccinia virus scaffold protein D13 trimer with N-terminal poly-histidine tag (half map 2). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vaccinia virus scaffolding protein D13 with N-terminal polyhistid...
| Entire | Name: Vaccinia virus scaffolding protein D13 with N-terminal polyhistidine tag in its trimeric state |
|---|---|
| Components |
|
-Supramolecule #1: Vaccinia virus scaffolding protein D13 with N-terminal polyhistid...
| Supramolecule | Name: Vaccinia virus scaffolding protein D13 with N-terminal polyhistidine tag in its trimeric state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinant D13 was expressed with N-terminal polyhistidine-tag using bacterial expression system. The protein was purified using metal affinity chromatography and size exclusion ...Details: Recombinant D13 was expressed with N-terminal polyhistidine-tag using bacterial expression system. The protein was purified using metal affinity chromatography and size exclusion chromatography. The final purified protein was trimeric. |
|---|---|
| Source (natural) | Organism:  Vaccinia virus WR Vaccinia virus WR |
| Molecular weight | Theoretical: 195 KDa |
-Macromolecule #1: Scaffold protein D13
| Macromolecule | Name: Scaffold protein D13 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vaccinia virus (strain Western Reserve) / Strain: Western Reserve Vaccinia virus (strain Western Reserve) / Strain: Western Reserve |
| Molecular weight | Theoretical: 64.16293 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHDYDI PTTENLYFQG AMNNTIINSL IGGDDSIKRS NVFAVDSQIP TLYMPQYISL SGVMTNDGPD NQAIASFEIR DQYITALNH LVLSLELPEV KGMGRFGYVP YVGYKCINHV SISSCNGVIW EIEGEELYNN CINNTIALKH SGYSSELNDI S IGLTPNDT ...String: HHHHHHDYDI PTTENLYFQG AMNNTIINSL IGGDDSIKRS NVFAVDSQIP TLYMPQYISL SGVMTNDGPD NQAIASFEIR DQYITALNH LVLSLELPEV KGMGRFGYVP YVGYKCINHV SISSCNGVIW EIEGEELYNN CINNTIALKH SGYSSELNDI S IGLTPNDT IKEPSTVYVY IKTPFDVEDT FSSLKLSDSK ITVTVTFNPV SDIVIRDSSF DFETFNKEFV YVPELSFIGY MV KNVQIKP SFIEKPRRVI GQINQPTATV TEVHAATSLS VYTKPYYGNT DNKFISYPGY SQDEKDYIDA YVSRLLDDLV IVS DGPPTG YPESAEIVEV PEDGIVSIQD ADVYVKIDNV PDNMSVYLHT NLLMFGTRKN SFIYNISKKF SAITGTYSDA TKRT IFAHI SHSINIIDTS IPVSLWTSQR NVYNGDNRSA ESKAKDLFIN DPFIKGIDFK NKTDIISRLE VRFGNDVLYS ENGPI SRIY NELLTKSNNG TRTLTFNFTP KIFFRPTTIT ANVSRGKDKL SVRVVYSTMD VNHPIYYVQK QLVVVCNDLY KVSYDQ GVS ITKIMG UniProtKB: Scaffold protein OPG125 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 microliter sample volume was loaded onto a holey grid with additional graphene oxide film. 10 sec waiting time, 5 sec blotting time and blot force 0, no delay time were applied before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1418 / Average exposure time: 31.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 155000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)