+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30639 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of the DEAH-box helicase Prp2 | |||||||||
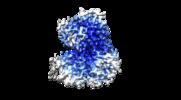
 Map data Map data | EM map of DEAH-box helicase Prp2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spliceosome / RNA splicing / Bact complex / DEAH-box ATPase/helicase / Prp2 / Spp2 / SPLICING | |||||||||
| Function / homology |  Function and homology information Function and homology informationsnoRNA splicing / generation of catalytic spliceosome for first transesterification step / U2-type catalytic step 1 spliceosome / ATP-dependent activity, acting on RNA / helicase activity / nucleic acid binding / RNA helicase activity / RNA helicase / ATP hydrolysis activity / RNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Bai R / Wan R | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2. Authors: Rui Bai / Ruixue Wan / Chuangye Yan / Qi Jia / Jianlin Lei / Yigong Shi /  Abstract: Spliceosome remodeling, executed by conserved adenosine triphosphatase (ATPase)/helicases including Prp2, enables precursor messenger RNA (pre-mRNA) splicing. However, the structural basis for the ...Spliceosome remodeling, executed by conserved adenosine triphosphatase (ATPase)/helicases including Prp2, enables precursor messenger RNA (pre-mRNA) splicing. However, the structural basis for the function of the ATPase/helicases remains poorly understood. Here, we report atomic structures of Prp2 in isolation, Prp2 complexed with its coactivator Spp2, and Prp2-loaded activated spliceosome and the results of structure-guided biochemical analysis. Prp2 weakly associates with the spliceosome and cannot function without Spp2, which stably associates with Prp2 and anchors on the spliceosome, thus tethering Prp2 to the activated spliceosome and allowing Prp2 to function. Pre-mRNA is loaded into a featured channel between the N and C halves of Prp2, where Leu from the N half and Arg from the C half prevent backward sliding of pre-mRNA toward its 5'-end. Adenosine 5'-triphosphate binding and hydrolysis trigger interdomain movement in Prp2, which drives unidirectional stepwise translocation of pre-mRNA toward its 3'-end. These conserved mechanisms explain the coupling of spliceosome remodeling to pre-mRNA splicing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30639.map.gz emd_30639.map.gz | 95.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30639-v30.xml emd-30639-v30.xml emd-30639.xml emd-30639.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30639.png emd_30639.png | 168.6 KB | ||
| Filedesc metadata |  emd-30639.cif.gz emd-30639.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30639 http://ftp.pdbj.org/pub/emdb/structures/EMD-30639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30639 | HTTPS FTP |
-Related structure data
| Related structure data |  7dcqMC  7dcoC  7dcpC  7dcrC  7dd3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30639.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30639.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of DEAH-box helicase Prp2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.6625 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DEAH-box ATPase/helicase Prp2
| Entire | Name: DEAH-box ATPase/helicase Prp2 |
|---|---|
| Components |
|
-Supramolecule #1: DEAH-box ATPase/helicase Prp2
| Supramolecule | Name: DEAH-box ATPase/helicase Prp2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 101 kDa/nm |
-Macromolecule #1: PRP2 isoform 1
| Macromolecule | Name: PRP2 isoform 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 99.947492 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSITSETGK RRVKRTYEVT RQNDNAVRIE PSSLGEEEDK EAKDKNSALQ LKRSRYDPNK VFSNTNQGPE KNNLKGEQLG SQKKSSKYD EKITSNNELT TKKGLLGDSE NETKYASSNS KFNVEVTHKI KNAKEIDKIN RQRMWEEQQL RNAMAGQSDH P DDITLEGS ...String: MSSITSETGK RRVKRTYEVT RQNDNAVRIE PSSLGEEEDK EAKDKNSALQ LKRSRYDPNK VFSNTNQGPE KNNLKGEQLG SQKKSSKYD EKITSNNELT TKKGLLGDSE NETKYASSNS KFNVEVTHKI KNAKEIDKIN RQRMWEEQQL RNAMAGQSDH P DDITLEGS DKYDYVFDTD AMIDYTNEED DLLPEEKLQY EARLAQALET EEKRILTIQE ARKLLPVHQY KDELLQEIKK NQ VLIIMGE TGSGKTTQLP QYLVEDGFTD QGKLQIAITQ PRRVAATSVA ARVADEMNVV LGKEVGYQIR FEDKTTPNKT VLK YMTDGM LLREFLTDSK LSKYSCIMID EAHERTLATD ILIGLLKDIL PQRPTLKLLI SSATMNAKKF SEFFDNCPIF NVPG RRYPV DIHYTLQPEA NYIHAAITTI FQIHTTQSLP GDILVFLTGQ EEIERTKTKL EEIMSKLGSR TKQMIITPIY ANLPQ EQQL KIFQPTPENC RKVVLATNIA ETSLTIDGIR YVIDPGFVKE NSYVPSTGMT QLLTVPCSRA SVDQRAGRAG RVGPGK CFR IFTKWSYLHE LELMPKPEIT RTNLSNTVLL LLSLGVTDLI KFPLMDKPSI PTLRKSLENL YILGALNSKG TITRLGK MM CEFPCEPEFA KVLYTAATHE QCQGVLEECL TIVSMLHETP SLFIGQKRDA AASVLSEVES DHILYLEIFN QWRNSKFS R SWCQDHKIQF KTMLRVRNIR NQLFRCSEKV GLVEKNDQAR MKIGNIAGYI NARITRCFIS GFPMNIVQLG PTGYQTMGR SSGGLNVSVH PTSILFVNHK EKAQRPSKYV LYQQLMLTSK EFIRDCLVIP KEEWLIDMVP QIFKDLIDDK TNRGRR UniProtKB: PRP2 isoform 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















