+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25691 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AKT1 K+ channel from A. thaliana in MSP2N2 lipid nanodisc | |||||||||
 Map data Map data | Sharpened electrostatic potential map from cryoSPARC non-uniform refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | K+ channel / ion channel / hyperpolarization-activated channel / voltage-gated channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationroot hair elongation / regulation of stomatal closure / response to water deprivation / inward rectifier potassium channel activity / potassium ion import across plasma membrane / monoatomic ion channel complex / response to salt stress / potassium ion transport / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Dickinson MS / Pourmal S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2022 Journal: Biochemistry / Year: 2022Title: Symmetry Reduction in a Hyperpolarization-Activated Homotetrameric Ion Channel. Authors: Miles Sasha Dickinson / Sergei Pourmal / Meghna Gupta / Maxine Bi / Robert M Stroud /  Abstract: Plants obtain nutrients from the soil via transmembrane transporters and channels in their root hairs, from which ions radially transport in toward the xylem for distribution across the plant body. ...Plants obtain nutrients from the soil via transmembrane transporters and channels in their root hairs, from which ions radially transport in toward the xylem for distribution across the plant body. We determined structures of the hyperpolarization-activated channel AKT1 from , which mediates K uptake from the soil into plant roots. These structures of AtAKT1 embedded in lipid nanodiscs show that the channel undergoes a reduction of C4 to C2 symmetry, possibly to regulate its electrical activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25691.map.gz emd_25691.map.gz | 290.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25691-v30.xml emd-25691-v30.xml emd-25691.xml emd-25691.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25691.png emd_25691.png | 164.6 KB | ||
| Filedesc metadata |  emd-25691.cif.gz emd-25691.cif.gz | 6.8 KB | ||
| Others |  emd_25691_additional_1.map.gz emd_25691_additional_1.map.gz emd_25691_additional_2.map.gz emd_25691_additional_2.map.gz emd_25691_half_map_1.map.gz emd_25691_half_map_1.map.gz emd_25691_half_map_2.map.gz emd_25691_half_map_2.map.gz | 270.8 MB 13.2 MB 285.5 MB 285.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25691 http://ftp.pdbj.org/pub/emdb/structures/EMD-25691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25691 | HTTPS FTP |
-Validation report
| Summary document |  emd_25691_validation.pdf.gz emd_25691_validation.pdf.gz | 822.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25691_full_validation.pdf.gz emd_25691_full_validation.pdf.gz | 821.7 KB | Display | |
| Data in XML |  emd_25691_validation.xml.gz emd_25691_validation.xml.gz | 17 KB | Display | |
| Data in CIF |  emd_25691_validation.cif.gz emd_25691_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25691 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25691 | HTTPS FTP |
-Related structure data
| Related structure data |  7t4xMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25691.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25691.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened electrostatic potential map from cryoSPARC non-uniform refinement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Density modification using DeepEmhancer. Inputs are two half...
| File | emd_25691_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modification using DeepEmhancer. Inputs are two half maps from cryoSPARC non-uniform refinement, using the "high resolution" protocol. | ||||||||||||
| Projections & Slices |
| ||||||||||||
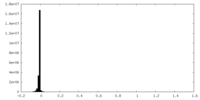
| Density Histograms |
-Additional map: Density modification using phenix.resolve cryo em. Input is two half...
| File | emd_25691_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modification using phenix.resolve_cryo_em. Input is two half maps from C4-symmetrized non-uniform refinement (cryoSPARC). | ||||||||||||
| Projections & Slices |
| ||||||||||||
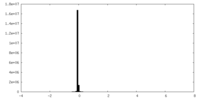
| Density Histograms |
-Half map: Half map 1 from cryoSPARC non-uniform refinement
| File | emd_25691_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from cryoSPARC non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 from cryoSPARC non-uniform refinement
| File | emd_25691_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from cryoSPARC non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotetramer of AKT1 subunits
| Entire | Name: Homotetramer of AKT1 subunits |
|---|---|
| Components |
|
-Supramolecule #1: Homotetramer of AKT1 subunits
| Supramolecule | Name: Homotetramer of AKT1 subunits / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 969.89 KDa |
-Macromolecule #1: Potassium channel AKT1
| Macromolecule | Name: Potassium channel AKT1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.109625 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRGGALLCGQ VQDEIEQLSR ESSHFSLSTG ILPSLGARSN RRVKLRRFVV SPYDHKYRIW EAFLVVLVVY TAWVSPFEFG FLRKPRPPL SITDNIVNAF FAIDIIMTFF VGYLDKSTYL IVDDRKQIAF KYLRSWFLLD LVSTIPSEAA MRISSQSYGL F NMLRLWRL ...String: MRGGALLCGQ VQDEIEQLSR ESSHFSLSTG ILPSLGARSN RRVKLRRFVV SPYDHKYRIW EAFLVVLVVY TAWVSPFEFG FLRKPRPPL SITDNIVNAF FAIDIIMTFF VGYLDKSTYL IVDDRKQIAF KYLRSWFLLD LVSTIPSEAA MRISSQSYGL F NMLRLWRL RRVGALFARL EKDRNFNYFW VRCAKLVCVT LFAVHCAACF YYLIAARNSN PAKTWIGANV ANFLEESLWM RY VTSMYWS ITTLTTVGYG DLHPVNTKEM IFDIFYMLFN LGLTAYLIGN MTNLVVHGTS RTRNFRDTIQ AASNFAHRNH LPP RLQDQM LAHLCLKYRT DSEGLQQQET LDALPKAIRS SISHFLFYSL MDKVYLFRGV SNDLLFQLVS EMKAEYFPPK EDVI LQNEA PTDFYILVNG TADLVDVDTG TESIVREVKA GDIIGEIGVL CYRPQLFTVR TKRLCQLLRM NRTTFLNIIQ ANVGD GTII MNNLLQHLKE MNDPVMTNVL LEIENMLARG KMDLPLNLCF AAIREDDLLL HQLLKRGLDP NESDNNGRTP LHIAAS KGT LNCVLLLLEY HADPNCRDAE GSVPLWEAMV EGHEKVVKVL LEHGSTIDAG DVGHFACTAA EQGNLKLLKE IVLHGGD VT RPRATGTSAL HTAVCEENIE MVKYLLEQGA DVNKQDMHGW TPRDLAEQQG HEDIKALFRE KLHERRVHIE TSSSVPIL K TGIRFLGRFT SEPNIRPASR EVSFRIRETR ARRKTNNFDN SLFGILANQS VPKNGLATVD EGRTGNPVRV TISCAEKDD IAGKLVLLPG SFKELLELGS NKFGIVATKV MNKDNNAEID DVDVIRDGDH LIFATDS UniProtKB: Potassium channel AKT1 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 13 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Homotetramer of AKT1 subunits in MSP2N2 lipid nanodisc |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 4660 / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)