+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24248 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Partial C. difficile TcdB and CSPG4 fragment | |||||||||
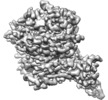
 Map data Map data | Partial C. difficile TcdB with CSPG4 (410-560) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TOXIN / TOXIN-Developmental Protein complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective CHST3 causes SEDCJD / Defective CHST14 causes EDS, musculocontractural type / Defective CHSY1 causes TPBS / DS-GAG biosynthesis / CS-GAG biosynthesis / CS/DS degradation / substrate-dependent cell migration / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD ...Defective CHST3 causes SEDCJD / Defective CHST14 causes EDS, musculocontractural type / Defective CHSY1 causes TPBS / DS-GAG biosynthesis / CS-GAG biosynthesis / CS/DS degradation / substrate-dependent cell migration / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / symbiont-mediated perturbation of host actin cytoskeleton via filamentous actin depolymerization / Glycosaminoglycan-protein linkage region biosynthesis / glial cell migration / tissue remodeling / ruffle assembly / glucosyltransferase activity / Differentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin / positive regulation of peptidyl-tyrosine phosphorylation / Transferases; Glycosyltransferases; Hexosyltransferases / host cell cytosol / platelet-derived growth factor receptor signaling pathway / lamellipodium membrane / coreceptor activity / ruffle / cysteine-type peptidase activity / lysosomal lumen / host cell endosome membrane / Golgi lumen / : / toxin activity / angiogenesis / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / positive regulation of MAPK cascade / intracellular signal transduction / apical plasma membrane / focal adhesion / lipid binding / protein kinase binding / host cell plasma membrane / cell surface / proteolysis / extracellular exosome / extracellular region / nucleoplasm / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Clostridioides difficile (bacteria) / Clostridioides difficile (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Jiang M / Zhang J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification Authors: Jiang M / Zhang J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24248.map.gz emd_24248.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24248-v30.xml emd-24248-v30.xml emd-24248.xml emd-24248.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24248.png emd_24248.png | 135.6 KB | ||
| Filedesc metadata |  emd-24248.cif.gz emd-24248.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24248 http://ftp.pdbj.org/pub/emdb/structures/EMD-24248 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24248 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24248 | HTTPS FTP |
-Related structure data
| Related structure data |  7n8xMC  7n95C  7n97C  7n9qC  7n9rC  7n9sC  7n9yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24248.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24248.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Partial C. difficile TcdB with CSPG4 (410-560) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The complex of TcdB and CSPG4 fragment
| Entire | Name: The complex of TcdB and CSPG4 fragment |
|---|---|
| Components |
|
-Supramolecule #1: The complex of TcdB and CSPG4 fragment
| Supramolecule | Name: The complex of TcdB and CSPG4 fragment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
| Molecular weight | Theoretical: 250 KDa |
-Supramolecule #2: Ternary structure of C-terminus of CSPG4 domain 1
| Supramolecule | Name: Ternary structure of C-terminus of CSPG4 domain 1 / type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Ternary structure of Clostridium difficile TcdB
| Supramolecule | Name: Ternary structure of Clostridium difficile TcdB / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
-Macromolecule #1: Chondroitin sulfate proteoglycan 4
| Macromolecule | Name: Chondroitin sulfate proteoglycan 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.565927 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LPEPCVPEPG LPPVFANFTQ LLTISPLVVA EGGTAWLEWR HVQPTLDLME AELRKSQVLF SVTRGARHGE LELDIPGAQA RKMFTLLDV VNRKARFIHD GSEDTSDQLV LEVSVTARVP MPSCLRRGQT YLLPIQVNPV N UniProtKB: Chondroitin sulfate proteoglycan 4 |
-Macromolecule #2: Toxin B
| Macromolecule | Name: Toxin B / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
| Molecular weight | Theoretical: 169.479141 KDa |
| Recombinant expression | Organism:  Bacillus megaterium NBRC 15308 = ATCC 14581 (bacteria) Bacillus megaterium NBRC 15308 = ATCC 14581 (bacteria) |
| Sequence | String: KIAFNSKGII NQGLISVKDS YCSNLIVKQI ENRYKILNNS LNPAISEDND FNTTTNTFID SIMAEANADN GRFMMELGKY LRVGFFPDV KTTINLSGPE AYAAAYQDLL MFKEGSMNIH LIEADLRNFE ISKTNISQST EQEMASLWSF DDARAKAQFE E YKRNYFEG ...String: KIAFNSKGII NQGLISVKDS YCSNLIVKQI ENRYKILNNS LNPAISEDND FNTTTNTFID SIMAEANADN GRFMMELGKY LRVGFFPDV KTTINLSGPE AYAAAYQDLL MFKEGSMNIH LIEADLRNFE ISKTNISQST EQEMASLWSF DDARAKAQFE E YKRNYFEG SLGEDDNLDF SQNIVVDKEY LLEKISSLAR SSERGYIHYI VQLQGDKISY EAACNLFAKT PYDSVLFQKN IE DSEIAYY YNPGDGEIQE IDKYKIPSII SDRPKIKLTF IGHGKDEFNT DIFAGFDVDS LSTEIEAAID LAKEDISPKS IEI NLLGCN MFSYSINVEE TYPGKLLLKV KDKISELMPS ISQDSIIVSA NQYEVRINSE GRRELLDHSG EWINKEESII KDIS SKEYI SFNPKENKIT VKSKNLPELS TLLQEIRNNS NSSDIELEEK VMLTECEINV ISNIDTQIVE ERIEEAKNLT SDSIN YIKD EFKLIESISD ALCDLKQQNE LEDSHFISFE DISETDEGFS IRFINKETGE SIFVETEKTI FSEYANHITE EISKIK GTI FDTVNGKLVK KVNLDTTHEV NTLNAAFFIQ SLIEYNSSKE SLSNLSVAMK VQVYAQLFST GLNTITDAAK VVELVST AL DETIDLLPTL SEGLPIIATI IDGVSLGAAI KELSETSDPL LRQEIEAKIG IMAVNLTTAT TAIITSSLGI ASGFSILL V PLAGISAGIP SLVNNELVLR DKATKVVDYF KHVSLVETEG VFTLLDDKIM MPQDDLVISE IDFNNNSIVL GKCEIWRME GGSGHTVTDD IDHFFSAPSI TYREPHLSIY DVLEVQKEEL DLSKDLMVLP NAPNRVFAWE TGWTPGLRSL ENDGTKLLDR IRDNYEGEF YWRYFAFIAD ALITTLKPRY EDTNIRINLD SNTRSFIVPI ITTEYIREKL SYSFYGSGGT YALSLSQYNM G INIELSES DVWIIDVDNV VRDVTIESDK IKKGDLIEGI LSTLSIEENK IILNSHEINF SGEVNGSNGF VSLTFSILEG IN AIIEVDL LSKSYKLLIS GELKILMLNS NHIQQKIDYI GFNSELQKNI PYSFVDSEGK ENGFINGSTK EGLFVSELPD VVL ISKVYM DDSKPSFGYY SNNLKDVKVI TKDNVNILTG YYLKDDIKIS LSLTLQDEKT IKLNSVHLDE SGVAEILKFM NRKG NTNTS DSLMSFLESM NIKSIFVNFL QSNIKFILDA NFIISGTTSI GQFEFICDEN DNIQPYFIKF NTLETNYTLY VGNRQ NMIV EPNYDLDDSG DISSTVINFS QKYLYGIDSC VNKVVISPNI YTDEINITPV YETNNTYPEV IVLDANYINE KINVNI NDL SIRYVWSNDG NDFILMSTSE ENKVSQVKIR FVNVFKDKTL ANKLSFNFSD KQDVPVSEII LSFTPSYYED GLIGYDL GL VSLYNEKFYI NNFGMMVSGL IYINDSLYYF KPPVNNLITG FVTVGDDKYY FNPINGGAAS I UniProtKB: Toxin B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7n8x: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















