[English] 日本語
 Yorodumi
Yorodumi- EMDB-23401: Cryo-EM structure of the B dENE construct complexed with a 28-mer... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23401 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) | ||||||||||||
 Map data Map data | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A). | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | RNA triple helix / RNA stability / poly(A) / SAXS and cryo-EM / RNA | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | ||||||||||||
 Authors Authors | Torabi S / Chen Y | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structural analyses of an RNA stability element interacting with poly(A). Authors: Seyed-Fakhreddin Torabi / Yen-Lin Chen / Kaiming Zhang / Jimin Wang / Suzanne J DeGregorio / Anand T Vaidya / Zhaoming Su / Suzette A Pabit / Wah Chiu / Lois Pollack / Joan A Steitz /   Abstract: Cis-acting RNA elements are crucial for the regulation of polyadenylated RNA stability. The element for nuclear expression (ENE) contains a U-rich internal loop flanked by short helices. An ENE ...Cis-acting RNA elements are crucial for the regulation of polyadenylated RNA stability. The element for nuclear expression (ENE) contains a U-rich internal loop flanked by short helices. An ENE stabilizes RNA by sequestering the poly(A) tail via formation of a triplex structure that inhibits a rapid deadenylation-dependent decay pathway. Structure-based bioinformatic studies identified numerous ENE-like elements in evolutionarily diverse genomes, including a subclass containing two ENE motifs separated by a short double-helical region (double ENEs [dENEs]). Here, the structure of a dENE derived from a rice transposable element (TWIFB1) before and after poly(A) binding (∼24 kDa and ∼33 kDa, respectively) is investigated. We combine biochemical structure probing, small angle X-ray scattering (SAXS), and cryo-electron microscopy (cryo-EM) to investigate the dENE structure and its local and global structural changes upon poly(A) binding. Our data reveal 1) the directionality of poly(A) binding to the dENE, and 2) that the dENE-poly(A) interaction involves a motif that protects the 3'-most seven adenylates of the poly(A). Furthermore, we demonstrate that the dENE does not undergo a dramatic global conformational change upon poly(A) binding. These findings are consistent with the recently solved crystal structure of a dENE+poly(A) complex [S.-F. Torabi , 371, eabe6523 (2021)]. Identification of additional modes of poly(A)-RNA interaction opens new venues for better understanding of poly(A) tail biology. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23401.map.gz emd_23401.map.gz | 21.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23401-v30.xml emd-23401-v30.xml emd-23401.xml emd-23401.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23401.png emd_23401.png | 31.3 KB | ||
| Filedesc metadata |  emd-23401.cif.gz emd-23401.cif.gz | 4.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23401 http://ftp.pdbj.org/pub/emdb/structures/EMD-23401 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23401 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23401 | HTTPS FTP |
-Validation report
| Summary document |  emd_23401_validation.pdf.gz emd_23401_validation.pdf.gz | 434.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23401_full_validation.pdf.gz emd_23401_full_validation.pdf.gz | 434.2 KB | Display | |
| Data in XML |  emd_23401_validation.xml.gz emd_23401_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_23401_validation.cif.gz emd_23401_validation.cif.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23401 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23401 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23401 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23401 | HTTPS FTP |
-Related structure data
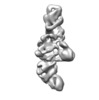
| Related structure data | 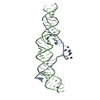 7ljyMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23401.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23401.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : dENE construct complexed with a 28-mer poly(A)
| Entire | Name: dENE construct complexed with a 28-mer poly(A) |
|---|---|
| Components |
|
-Supramolecule #1: dENE construct complexed with a 28-mer poly(A)
| Supramolecule | Name: dENE construct complexed with a 28-mer poly(A) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33 KDa |
-Macromolecule #1: B dENE construct
| Macromolecule | Name: B dENE construct / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.242186 KDa |
| Sequence | String: GGGUACUCUU UUCUUUGUCA UGGUUUUCUC AGGCGAAAGU CUGAGUUUUU ACAUGACAAA GUUUUUAACG AGGCCC |
-Macromolecule #2: poly(A)
| Macromolecule | Name: poly(A) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.172806 KDa |
| Sequence | String: AAAAAAAAAA AAAAAAAAAA AAAAAAAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 5.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15.0) / Number images used: 283486 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7ljy: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















