+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23321 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phage Qbeta T=4 particle | |||||||||||||||
 Map data Map data | Qbeta T=4 virus-like particle | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Bacteriophage / Virus / Qbeta / VIRUS LIKE PARTICLE | |||||||||||||||
| Function / homology | Levivirus coat protein / Levivirus coat protein / Bacteriophage RNA-type, capsid / T=3 icosahedral viral capsid / translation repressor activity / structural molecule activity / RNA binding / Capsid protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Escherichia phage Qbeta (virus) / Escherichia phage Qbeta (virus) /  Escherichia virus Qbeta Escherichia virus Qbeta | |||||||||||||||
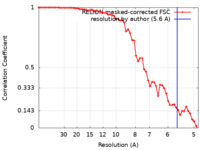
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||||||||
 Authors Authors | Chang JY / Zhang J | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles. Authors: Jeng-Yih Chang / Karl V Gorzelnik / Jirapat Thongchol / Junjie Zhang /  Abstract: The coat proteins (CPs) of single-stranded RNA bacteriophages (ssRNA phages) directly assemble around the genomic RNA (gRNA) to form a near-icosahedral capsid with a single maturation protein (Mat) ...The coat proteins (CPs) of single-stranded RNA bacteriophages (ssRNA phages) directly assemble around the genomic RNA (gRNA) to form a near-icosahedral capsid with a single maturation protein (Mat) that binds the gRNA and interacts with the retractile pilus during infection of the host. Understanding the assembly of ssRNA phages is essential for their use in biotechnology, such as RNA protection and delivery. Here, we present the complete gRNA model of the ssRNA phage Qβ, revealing that the 3' untranslated region binds to the Mat and the 4127 nucleotides fold domain-by-domain, and is connected through long-range RNA-RNA interactions, such as kissing loops. Thirty-three operator-like RNA stem-loops are located and primarily interact with the asymmetric A/B CP-dimers, suggesting a pathway for the assembly of the virions. Additionally, we have discovered various forms of the virus-like particles (VLPs), including the canonical = 3 icosahedral, larger = 4 icosahedral, prolate, oblate forms, and a small prolate form elongated along the 3-fold axis. These particles are all produced during a normal infection, as well as when overexpressing the CPs. When overexpressing the shorter RNA fragments encoding only the CPs, we observed an increased percentage of the smaller VLPs, which may be sufficient to encapsidate a shorter RNA. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23321.map.gz emd_23321.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23321-v30.xml emd-23321-v30.xml emd-23321.xml emd-23321.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23321_fsc.xml emd_23321_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_23321.png emd_23321.png | 286.6 KB | ||
| Filedesc metadata |  emd-23321.cif.gz emd-23321.cif.gz | 4.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23321 http://ftp.pdbj.org/pub/emdb/structures/EMD-23321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23321 | HTTPS FTP |
-Validation report
| Summary document |  emd_23321_validation.pdf.gz emd_23321_validation.pdf.gz | 527.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23321_full_validation.pdf.gz emd_23321_full_validation.pdf.gz | 526.7 KB | Display | |
| Data in XML |  emd_23321_validation.xml.gz emd_23321_validation.xml.gz | 8.8 KB | Display | |
| Data in CIF |  emd_23321_validation.cif.gz emd_23321_validation.cif.gz | 11.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23321 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23321 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23321 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23321 | HTTPS FTP |
-Related structure data
| Related structure data |  7lgeMC  7lgfC  7lggC  7lghC  7lhdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23321.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23321.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Qbeta T=4 virus-like particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.432 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Escherichia virus Qbeta
| Entire | Name:  Escherichia virus Qbeta Escherichia virus Qbeta |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus Qbeta
| Supramolecule | Name: Escherichia virus Qbeta / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 39803 / Sci species name: Escherichia virus Qbeta / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Qbeta (virus) Escherichia phage Qbeta (virus) |
| Molecular weight | Theoretical: 14.268071 KDa |
| Sequence | String: MAKLETVTLG NIGKDGKQTL VLNPRGVNPT NGVASLSQAG AVPALEKRVT VSVSQPSRNR KNYKVQVKIQ NPTACTANGS CDPSVTRQA YADVTFSFTQ YSTDEERAFV RTELAALLAS PLLIDAIDQL NPAY UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | JEOL 3200FSC |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TECNAI F20 |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)