[English] 日本語
 Yorodumi
Yorodumi- EMDB-22252: CRYOEM STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22252 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRYOEM STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE ZMP1 IN OPEN STATE | |||||||||
 Map data Map data | ZINC METALLOPROTEASE ZMP1 IN OPEN STATE | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Open state / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmetalloendopeptidase activity / protein processing / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||
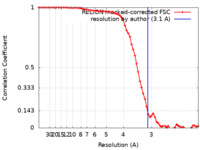
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Liang WG / Zhao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Structural analysis of Mycobacterium tuberculosis M13 metalloprotease Zmp1 open states. Authors: Wenguang G Liang / Jordan M Mancl / Minglei Zhao / Wei-Jen Tang /  Abstract: Zinc metalloprotease 1 (Zmp1), a Mycobacterium tuberculosis 75 kDa secreted enzyme, mediates key stages of tuberculosis disease progression. The biological activity of Zmp1 presumably stems from its ...Zinc metalloprotease 1 (Zmp1), a Mycobacterium tuberculosis 75 kDa secreted enzyme, mediates key stages of tuberculosis disease progression. The biological activity of Zmp1 presumably stems from its ability to degrade bacterium- and/or host-derived peptides. The crystal structures of Zmp1 and related M13 metalloproteases, such as neprilysin and endothelin-converting enzyme-1 were determined only in the closed conformation, which cannot capture substrates or release proteolytic products. Thus, the mechanisms of substrate binding and selectivity remain elusive. Here we report two open-state cryo-EM structures of Zmp1, revealed by our SAXS analysis to be the dominant states in solution. Our structural analyses reveal how ligand binding induces a conformational switch in four linker regions to drive the rigid body motion of the D1 and D2 domains, which form the sizable catalytic chamber. Furthermore, they offer insights into the catalytic cycle and mechanism of substrate recognition of M13 metalloproteases for future therapeutic innovations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22252.map.gz emd_22252.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22252-v30.xml emd-22252-v30.xml emd-22252.xml emd-22252.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22252_fsc.xml emd_22252_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22252.png emd_22252.png | 110.6 KB | ||
| Masks |  emd_22252_msk_1.map emd_22252_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22252.cif.gz emd-22252.cif.gz | 6.7 KB | ||
| Others |  emd_22252_additional_1.map.gz emd_22252_additional_1.map.gz emd_22252_half_map_1.map.gz emd_22252_half_map_1.map.gz emd_22252_half_map_2.map.gz emd_22252_half_map_2.map.gz | 25.3 MB 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22252 http://ftp.pdbj.org/pub/emdb/structures/EMD-22252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22252 | HTTPS FTP |
-Related structure data
| Related structure data |  6xlyMC  7k1vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22252.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22252.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZINC METALLOPROTEASE ZMP1 IN OPEN STATE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
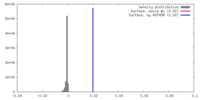
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22252_msk_1.map emd_22252_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
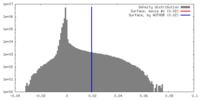
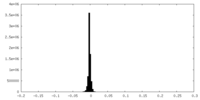
| Density Histograms |
-Additional map: sharpen map
| File | emd_22252_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
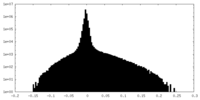
| Density Histograms |
-Half map: ZINC METALLOPROTEASE ZMP1 IN OPEN STATE
| File | emd_22252_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZINC METALLOPROTEASE ZMP1 IN OPEN STATE | ||||||||||||
| Projections & Slices |
| ||||||||||||
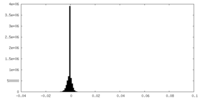
| Density Histograms |
-Half map: ZINC METALLOPROTEASE ZMP1 IN OPEN STATE
| File | emd_22252_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZINC METALLOPROTEASE ZMP1 IN OPEN STATE | ||||||||||||
| Projections & Slices |
| ||||||||||||
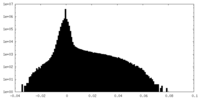
| Density Histograms |
- Sample components
Sample components
-Entire : MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE
| Entire | Name: MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE |
|---|---|
| Components |
|
-Supramolecule #1: MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE
| Supramolecule | Name: MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
-Macromolecule #1: Probable zinc metalloprotease Zmp1
| Macromolecule | Name: Probable zinc metalloprotease Zmp1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 78.132953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDHPFTMTLA IPSGIDLSHI DADARPQDDL FGHVNGRWLA EHEIPADRAT DGAFRSLFD RAETQVRDLI IQASQAGAAV GTDAQRIGDL YASFLDEEAV ERAGVQPLHD ELATIDSAAD ATELAAALGT L QRAGVGGG ...String: MRGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDHPFTMTLA IPSGIDLSHI DADARPQDDL FGHVNGRWLA EHEIPADRAT DGAFRSLFD RAETQVRDLI IQASQAGAAV GTDAQRIGDL YASFLDEEAV ERAGVQPLHD ELATIDSAAD ATELAAALGT L QRAGVGGG IGVYVDTDSK DSTRYLVHFT QSGIGLPDES YYRDEQHAAV LAAYPGHIAR MFGLVYGGES RDHAKTADRI VA LETKLAD AHWDVVKRRD ADLGYNLRTF AQLQTEGAGF DWVSWVTALG SAPDAMTELV VRQPDYLVTF ASLWASVNVE DWK CWARWR LIRARAPWLT RALVAEDFEF YGRTLTGAQQ LRDRWKRGVS LVENLMGDAV GKLYVQRHFP PDAKSRIDTL VDNL QEAYR ISISELDWMT PQTRQRALAK LNKFTAKVGY PIKWRDYSKL AIDRDDLYGN VQRGYAVNHD RELAKLFGPV DRDEW FMTP QTVNAYYNPG MNEIVFPAAI LQPPFFDPQA DEAANYGGIG AVIGHEIGHG FDDQGAKYDG DGNLVDWWTD DDRTEF AAR TKALIEQYHA YTPRDLVDHP GPPHVQGAFT IGENIGDLGG LSIALLAYQL SLNGNPAPVI DGLTGMQRVF FGWAQIW RT KSRAAEAIRR LAVDPHSPPE FRCNGVVRNV DAFYQAFDVT EDDALFLDPQ RRVRIWN UniProtKB: Probable zinc metalloprotease Zmp1 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 6.8 Details: 20 mM HEPES (pH 6.8), 150 mM NaCl, 0.5 mM bMe, 20 mM Trimethylamine N-oxide dihydrate |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 308 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 5.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)