[English] 日本語
 Yorodumi
Yorodumi- EMDB-21870: Molecular basis for the ATPase-powered substrate translocation by... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21870 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | ||||||||||||
 Map data Map data | Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Lon AAA+ / protease / cryo-EM / dual pore-loops / MOTOR PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase La / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / cellular response to heat / sequence-specific DNA binding / serine-type endopeptidase activity / ATP hydrolysis activity / ATP binding / metal ion binding / identical protein binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Meiothermus taiwanensis (bacteria) / Meiothermus taiwanensis (bacteria) /  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Zhang K / Li S | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2021 Journal: J Biol Chem / Year: 2021Title: Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease. Authors: Shanshan Li / Kan-Yen Hsieh / Shih-Chieh Su / Grigore D Pintilie / Kaiming Zhang / Chung-I Chang /    Abstract: The Lon AAA+ (adenosine triphosphatases associated with diverse cellular activities) protease (LonA) converts ATP-fuelled conformational changes into sufficient mechanical force to drive ...The Lon AAA+ (adenosine triphosphatases associated with diverse cellular activities) protease (LonA) converts ATP-fuelled conformational changes into sufficient mechanical force to drive translocation of a substrate into a hexameric proteolytic chamber. To understand the structural basis for the substrate translocation process, we determined the cryo-electron microscopy (cryo-EM) structure of Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state at 3.6 Å resolution. Our data indicate that substrate interactions are mediated by the dual pore loops of the ATPase domains, organized in spiral staircase arrangement from four consecutive protomers in different ATP-binding and hydrolysis states. However, a closed AAA+ ring is maintained by two disengaged ADP-bound protomers transiting between the lowest and highest position. This structure reveals a processive rotary translocation mechanism mediated by LonA-specific nucleotide-dependent allosteric coordination among the ATPase domains, which is induced by substrate binding. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21870.map.gz emd_21870.map.gz | 136.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21870-v30.xml emd-21870-v30.xml emd-21870.xml emd-21870.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21870.png emd_21870.png | 212.7 KB | ||
| Filedesc metadata |  emd-21870.cif.gz emd-21870.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21870 http://ftp.pdbj.org/pub/emdb/structures/EMD-21870 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21870 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21870 | HTTPS FTP |
-Validation report
| Summary document |  emd_21870_validation.pdf.gz emd_21870_validation.pdf.gz | 561.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21870_full_validation.pdf.gz emd_21870_full_validation.pdf.gz | 560.7 KB | Display | |
| Data in XML |  emd_21870_validation.xml.gz emd_21870_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_21870_validation.cif.gz emd_21870_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21870 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21870 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21870 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21870 | HTTPS FTP |
-Related structure data
| Related structure data |  6wqhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21870.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21870.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state
| Entire | Name: Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state |
|---|---|
| Components |
|
-Supramolecule #1: Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state
| Supramolecule | Name: Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 360 KDa |
-Supramolecule #2: Lon protease
| Supramolecule | Name: Lon protease / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
-Supramolecule #3: Ig2 substrate
| Supramolecule | Name: Ig2 substrate / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Lon protease
| Macromolecule | Name: Lon protease / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
| Molecular weight | Theoretical: 88.701766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRLELPVIPL RNTVILPHTT TPVDVGRAKS KRAVEEAMGA DRLIFLVAQR DPEVDDPAPD DLYTWGVQAV VKQAMRLPDG TLQVMVEAR ARAFQVTDYI PGPYLRARGE VFSEIFPIDE AVVRVLVEEL KEAFEKYVAN HKSLRLDRYQ LEAVKGTSDP A MLADTIAY ...String: MRLELPVIPL RNTVILPHTT TPVDVGRAKS KRAVEEAMGA DRLIFLVAQR DPEVDDPAPD DLYTWGVQAV VKQAMRLPDG TLQVMVEAR ARAFQVTDYI PGPYLRARGE VFSEIFPIDE AVVRVLVEEL KEAFEKYVAN HKSLRLDRYQ LEAVKGTSDP A MLADTIAY HATWTVAEKQ EILELTDLEA RLKKVLGLLS RDLERFELDK RVAQRVKEQM DTNQREYYLR EQMKAIQKEL GG EDGLSDL EALRKKIEEV GMPEAVKTKA LKELDRLERM QQGSPEATVA RTYLDWLTEV PWSKADPEVL DINHTRQVLD EDH YGLKDV KERILEYLAV RQLTQGLDVR NKAPILVLVG PPGVGKTSLG RSIARSMNRK FHRISLGGVR DEAEIRGHRR TYIG AMPGK LIHAMKQVGV INPVILLDEI DKMSSDWRGD PASAMLEVLD PEQNNTFTDH YLDVPYDLSK VFFITTANTL QTIPR PLLD RMEVIEIPGY TNMEKQAIAR QYLWPKQVRE SGMEGRIEVT DAAILRVISE YTREAGVRGL ERELGKIARK GAKFWL EGA WEGLRTIDAS DIPTYLGIPR YRPDKAETEP QVGTAQGLAW TPVGGTLLTI EVAAVPGSGK LSLTGQLGEV MKESAQA AL TYLRAHTQDY GLPEDFYNKV DLHVHVPDGA TPKDGPSAGI TMATAIASAL SRRPARMDIA MTGEVSLRGK VMPIGGVK E KLLAAHQAGI HKIVLPKDNE AQLEELPKEV LEGLEIKLVE DVGEVLEYLL LPEPTMPPVV QPSDNRQQPG AGA UniProtKB: Lon protease |
-Macromolecule #2: Ig2 substrate
| Macromolecule | Name: Ig2 substrate / type: protein_or_peptide / ID: 2 / Details: Unfolded substate / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 954.168 Da |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 3 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #4: N-[(1R)-1-(dihydroxyboranyl)-2-phenylethyl]-Nalpha-(pyrazin-2-ylc...
| Macromolecule | Name: N-[(1R)-1-(dihydroxyboranyl)-2-phenylethyl]-Nalpha-(pyrazin-2-ylcarbonyl)-L-phenylalaninamide type: ligand / ID: 4 / Number of copies: 6 / Formula: 4KZ |
|---|---|
| Molecular weight | Theoretical: 418.253 Da |
| Chemical component information | 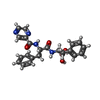 ChemComp-4KZ: |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1800 / Average exposure time: 6.0 sec. / Average electron dose: 8.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0.2) / Number images used: 23487 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















