+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21437 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of ABCG2 bound to imatinib | |||||||||
 Map data Map data | Cryo-EM map of ABCG2 bound to imatinib | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABCG2 / ABC transporter / imatinib / TRANSLOCASE-TRANSLOCASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationbiotin transmembrane transporter activity / biotin transport / riboflavin transport / riboflavin transmembrane transporter activity / sphingolipid transporter activity / renal urate salt excretion / Abacavir transmembrane transport / urate metabolic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis ...biotin transmembrane transporter activity / biotin transport / riboflavin transport / riboflavin transmembrane transporter activity / sphingolipid transporter activity / renal urate salt excretion / Abacavir transmembrane transport / urate metabolic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / urate transmembrane transporter activity / external side of apical plasma membrane / xenobiotic transport across blood-brain barrier / organic anion transport / transepithelial transport / : / Ciprofloxacin ADME / Paracetamol ADME / export across plasma membrane / NFE2L2 regulating MDR associated enzymes / Differentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin / ABC-type xenobiotic transporter / Heme biosynthesis / cellular detoxification / ABC-type xenobiotic transporter activity / Heme degradation / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / transport across blood-brain barrier / brush border membrane / Iron uptake and transport / transmembrane transport / mitochondrial membrane / apical plasma membrane / membrane raft / protein homodimerization activity / ATP hydrolysis activity / nucleoplasm / ATP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
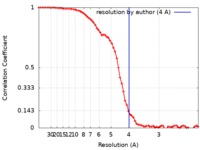
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Orlando BJ / Liao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: ABCG2 transports anticancer drugs via a closed-to-open switch. Authors: Benjamin J Orlando / Maofu Liao /  Abstract: ABCG2 is an ABC transporter that extrudes a variety of compounds from cells, and presents an obstacle in treating chemotherapy-resistant cancers. Despite recent structural insights, no anticancer ...ABCG2 is an ABC transporter that extrudes a variety of compounds from cells, and presents an obstacle in treating chemotherapy-resistant cancers. Despite recent structural insights, no anticancer drug bound to ABCG2 has been resolved, and the mechanisms of multidrug transport remain obscure. Such a gap of knowledge limits the development of novel compounds that block or evade this critical molecular pump. Here we present single-particle cryo-EM studies of ABCG2 in the apo state, and bound to the three structurally distinct chemotherapeutics. Without the binding of conformation-selective antibody fragments or inhibitors, the resting ABCG2 adopts a closed conformation. Our cryo-EM, biochemical, and functional analyses reveal the binding mode of three chemotherapeutic compounds, demonstrate how these molecules open the closed conformation of the transporter, and establish that imatinib is particularly effective in stabilizing the inward facing conformation of ABCG2. Together these studies reveal the previously unrecognized conformational cycle of ABCG2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21437.map.gz emd_21437.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21437-v30.xml emd-21437-v30.xml emd-21437.xml emd-21437.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21437_fsc.xml emd_21437_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_21437.png emd_21437.png | 124.2 KB | ||
| Filedesc metadata |  emd-21437.cif.gz emd-21437.cif.gz | 6.7 KB | ||
| Others |  emd_21437_additional.map.gz emd_21437_additional.map.gz | 20.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21437 http://ftp.pdbj.org/pub/emdb/structures/EMD-21437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21437 | HTTPS FTP |
-Related structure data
| Related structure data |  6vxhMC  6vxfC  6vxiC  6vxjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21437.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21437.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of ABCG2 bound to imatinib | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1333 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unfiltered and unsharpened cryo-EM map of ABCG2 bound to imatinib
| File | emd_21437_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered and unsharpened cryo-EM map of ABCG2 bound to imatinib | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ABCG2 reconstituted in lipid nanodisc
| Entire | Name: ABCG2 reconstituted in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: ABCG2 reconstituted in lipid nanodisc
| Supramolecule | Name: ABCG2 reconstituted in lipid nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Broad substrate specificity ATP-binding cassette transporter ABCG2
| Macromolecule | Name: Broad substrate specificity ATP-binding cassette transporter ABCG2 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type xenobiotic transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.385852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSSNVEVFI PVSQGNTNGF PATASNDLKA FTEGAVLSFH NICYRVKLKS GFLPCRKPVE KEILSNINGI MKPGLNAILG PTGGGKSSL LDVLAARKDP SGLSGDVLIN GAPRPANFKC NSGYVVQDDV VMGTLTVREN LQFSAALRLA TTMTNHEKNE R INRVIQEL ...String: MSSSNVEVFI PVSQGNTNGF PATASNDLKA FTEGAVLSFH NICYRVKLKS GFLPCRKPVE KEILSNINGI MKPGLNAILG PTGGGKSSL LDVLAARKDP SGLSGDVLIN GAPRPANFKC NSGYVVQDDV VMGTLTVREN LQFSAALRLA TTMTNHEKNE R INRVIQEL GLDKVADSKV GTQFIRGVSG GERKRTSIGM ELITDPSILF LDEPTTGLDS STANAVLLLL KRMSKQGRTI IF SIHQPRY SIFKLFDSLT LLASGRLMFH GPAQEALGYF ESAGYHCEAY NNPADFFLDI INGDSTAVAL NREEDFKATE IIE PSKQDK PLIEKLAEIY VNSSFYKETK AELHQLSGGE KKKKITVFKE ISYTTSFCHQ LRWVSKRSFK NLLGNPQASI AQII VTVVL GLVIGAIYFG LKNDSTGIQN RAGVLFFLTT NQCFSSVSAV ELFVVEKKLF IHEYISGYYR VSSYFLGKLL SDLLP MRML PSIIFTCIVY FMLGLKPKAD AFFVMMFTLM MVAYSASSMA LAIAAGQSVV SVATLLMTIC FVFMMIFSGL LVNLTT IAS WLSWLQYFSI PRYGFTALQH NEFLGQNFCP GLNATGNNPC NYATCTGEEY LVKQGIDLSP WGLWKNHVAL ACMIVIF LT IAYLKLLFLK KYS UniProtKB: Broad substrate specificity ATP-binding cassette transporter ABCG2 |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #3: 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-P...
| Macromolecule | Name: 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE type: ligand / ID: 3 / Number of copies: 1 / Formula: STI |
|---|---|
| Molecular weight | Theoretical: 493.603 Da |
| Chemical component information | 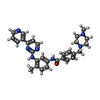 ChemComp-STI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 92 % / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)