[English] 日本語
 Yorodumi
Yorodumi- EMDB-20614: Structure of the Vesicular Stomatitis Virus L Protein in Complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20614 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | |||||||||
 Map data Map data | filtered, sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vesicular stomatitis virus / RNA-dependent RNA polymerase / L protein / P protein / phosphoprotein / single particle analysis / transcription / replication / virus / viral / RdRp / PRNTase / nonsegmented negative-sense RNA viruses / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationGDP polyribonucleotidyltransferase / RNA folding chaperone / negative stranded viral RNA replication / phosphorylation / viral transcription / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / viral genome replication / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity ...GDP polyribonucleotidyltransferase / RNA folding chaperone / negative stranded viral RNA replication / phosphorylation / viral transcription / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / viral genome replication / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / GTPase activity / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Vesicular stomatitis virus / Vesicular stomatitis virus /  Vesicular stomatitis Indiana virus (strain San Juan) Vesicular stomatitis Indiana virus (strain San Juan) | |||||||||
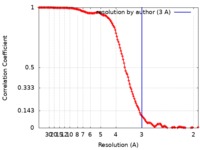
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Jenni S / Bloyet LM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor. Authors: Simon Jenni / Louis-Marie Bloyet / Ruben Diaz-Avalos / Bo Liang / Sean P J Whelan / Nikolaus Grigorieff / Stephen C Harrison /  Abstract: The large (L) proteins of non-segmented, negative-strand RNA viruses are multifunctional enzymes that produce capped, methylated, and polyadenylated mRNA and replicate the viral genome. A ...The large (L) proteins of non-segmented, negative-strand RNA viruses are multifunctional enzymes that produce capped, methylated, and polyadenylated mRNA and replicate the viral genome. A phosphoprotein (P), required for efficient RNA-dependent RNA polymerization from the viral ribonucleoprotein (RNP) template, regulates the function and conformation of the L protein. We report the structure of vesicular stomatitis virus L in complex with its P cofactor determined by electron cryomicroscopy at 3.0 Å resolution, enabling us to visualize bound segments of P. The contacts of three P segments with multiple L domains show how P induces a closed, compact, initiation-competent conformation. Binding of P to L positions its N-terminal domain adjacent to a putative RNA exit channel for efficient encapsidation of newly synthesized genomes with the nucleoprotein and orients its C-terminal domain to interact with an RNP template. The model shows that a conserved tryptophan in the priming loop can support the initiating 5' nucleotide. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20614.map.gz emd_20614.map.gz | 226.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20614-v30.xml emd-20614-v30.xml emd-20614.xml emd-20614.xml | 27.5 KB 27.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20614_fsc.xml emd_20614_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_20614.png emd_20614.png | 169.6 KB | ||
| Masks |  emd_20614_msk_1.map emd_20614_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20614.cif.gz emd-20614.cif.gz | 7.9 KB | ||
| Others |  emd_20614_additional_1.map.gz emd_20614_additional_1.map.gz emd_20614_additional_2.map.gz emd_20614_additional_2.map.gz emd_20614_additional_3.map.gz emd_20614_additional_3.map.gz emd_20614_half_map_1.map.gz emd_20614_half_map_1.map.gz emd_20614_half_map_2.map.gz emd_20614_half_map_2.map.gz | 226.7 MB 963.3 MB 33.1 MB 8.8 MB 8.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20614 http://ftp.pdbj.org/pub/emdb/structures/EMD-20614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20614 | HTTPS FTP |
-Related structure data
| Related structure data |  6u1xMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20614.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20614.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered, sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20614_msk_1.map emd_20614_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: filtered
| File | emd_20614_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: filtered, sharpened, calculated on fine grid
| File | emd_20614_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered, sharpened, calculated on fine grid | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: filtered, sharpened, calculated on fine grid, boxed
| File | emd_20614_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered, sharpened, calculated on fine grid, boxed | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map1, unmodified
| File | emd_20614_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1, unmodified | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map2, unmodified
| File | emd_20614_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2, unmodified | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VSV L:P
| Entire | Name: VSV L:P |
|---|---|
| Components |
|
-Supramolecule #1: VSV L:P
| Supramolecule | Name: VSV L:P / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: L protein (full-length) in complex with P protein (35-106) |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis virus Vesicular stomatitis virus |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis Indiana virus (strain San Juan) Vesicular stomatitis Indiana virus (strain San Juan)Strain: San Juan |
| Molecular weight | Theoretical: 241.313859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEVHDFETDE FNDFNEDDYA TREFLNPDER MTYLNHADYN LNSPLISDDI DNLIRKFNSL PIPSMWDSKN WDGVLEMLTS CQANPISTS QMHKWMGSWL MSDNHDASQG YSFLHEVDKE AEITFDVVET FIRGWGNKPI EYIKKERWTD SFKILAYLCQ K FLDLHKLT ...String: MEVHDFETDE FNDFNEDDYA TREFLNPDER MTYLNHADYN LNSPLISDDI DNLIRKFNSL PIPSMWDSKN WDGVLEMLTS CQANPISTS QMHKWMGSWL MSDNHDASQG YSFLHEVDKE AEITFDVVET FIRGWGNKPI EYIKKERWTD SFKILAYLCQ K FLDLHKLT LILNAVSEVE LLNLARTFKG KVRRSSHGTN ICRIRVPSLG PTFISEGWAY FKKLDILMDR NFLLMVKDVI IG RMQTVLS MVCRIDNLFS EQDIFSLLNI YRIGDKIVER QGNFSYDLIK MVEPICNLKL MKLARESRPL VPQFPHFENH IKT SVDEGA KIDRGIRFLH DQIMSVKTVD LTLVIYGSFR HWGHPFIDYY TGLEKLHSQV TMKKDIDVSY AKALASDLAR IVLF QQFND HKKWFVNGDL LPHDHPFKSH VKENTWPTAA QVQDFGDKWH ELPLIKCFEI PDLLDPSIIY SDKSHSMNRS EVLKH VRMN PNTPIPSKKV LQTMLDTKAT NWKEFLKEID EKGLDDDDLI IGLKGKEREL KLAGRFFSLM SWKLREYFVI TEYLIK THF VPMFKGLTMA DDLTAVIKKM LDSSSGQGLK SYEAICIANH IDYEKWNNHQ RKLSNGPVFR VMGQFLGYPS LIERTHE FF EKSLIYYNGR PDLMRVHNNT LINSTSQRVC WQGQEGGLEG LRQKGWTILN LLVIQREAKI RNTAVKVLAQ GDNQVICT Q YKTKKSRNVV ELQGALNQMV SNNEKIMTAI KIGTGKLGLL INDDETMQSA DYLNYGKIPI FRGVIRGLET KRWSRVTCV TNDQIPTCAN IMSSVSTNAL TVAHFAENPI NAMIQYNYFG TFARLLLMMH DPALRQSLYE VQDKIPGLHS STFKYAMLYL DPSIGGVSG MSLSRFLIRA FPDPVTESLS FWRFIHVHAR SEHLKEMSAV FGNPEIAKFR ITHIDKLVED PTSLNIAMGM S PANLLKTE VKKCLIESRQ TIRNQVIKDA TIYLYHEEDR LRSFLWSINP LFPRFLSEFK SGTFLGVADG LISLFQNSRT IR NSFKKKY HRELDDLIVR SEVSSLTHLG KLHLRRGSCK MWTCSATHAD TLRYKSWGRT VIGTTVPHPL EMLGPQHRKE TPC APCNTS GFNYVSVHCP DGIHDVFSSR GPLPAYLGSK TSESTSILQP WERESKVPLI KRATRLRDAI SWFVEPDSKL AMTI LSNIH SLTGEEWTKR QHGFKRTGSA LHRFSTSRMS HGGFASQSTA ALTRLMATTD TMRDLGDQNF DFLFQATLLY AQITT TVAR DGWITSCTDH YHIACKSCLR PIEEITLDSS MDYTPPDVSH VLKTWRNGEG SWGQEIKQIY PLEGNWKNLA PAEQSY QVG RCIGFLYGDL AYRKSTHAED SSLFPLSIQG RIRGRGFLKG LLDGLMRASC CQVIHRRSLA HLKRPANAVY GGLIYLI DK LSVSPPFLSL TRSGPIRDEL ETIPHKIPTS YPTSNRDMGV IVRNYFKYQC RLIEKGKYRS HYSQLWLFSD VLSIDFIG P FSISTTLLQI LYKPFLSGKD KNELRELANL SSLLRSGEGW EDIHVKFFTK DILLCPEEIR HACKFGIAKD NNKDMSYPP WGRESRGTIT TIPVYYTTTP YPKMLEMPPR IQNPLLSGIR LGQLPTGAHY KIRSILHGMG IHYRDFLSCG DGSGGMTAAL LRENVHSRG IFNSLLELSG SVMRGASPEP PSALETLGGD KSRCVNGETC WEYPSDLCDP RTWDYFLRLK AGLGLQIDLI V MDMEVRDS STSLKIETNV RNYVHRILDE QGVLIYKTYG TYICESEKNA VTILGPMFKT VDLVQTEFSS SQTSEVYMVC KG LKKLIDE PNPDWSSINE SWKNLYAFQS SEQEFARAKK VSTYFTLTGI PSQFIPDPFV NIETMLQIFG VPTGVSHAAA LKS SDRPAD LLTISLFYMA IISYYNINHI RVGPIPPNPP SDGIAQNVGI AITGISFWLS LMEKDIPLYQ QCLAVIQQSF PIRW EAVSV KGGYKQKWST RGDGLPKDTR ISDSLAPIGN WIRSLELVRN QVRLNPFNEI LFNQLCRTVD NHLKWSNLRR NTGMI EWIN RRISKEDRSI LMLKSDLHEE NSWRD UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis Indiana virus (strain San Juan) Vesicular stomatitis Indiana virus (strain San Juan)Strain: San Juan |
| Molecular weight | Theoretical: 29.941982 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDNLTKVREY LKSYSRLDQA VGEIDEIEAQ RAEKSNYELF QEDGVEEHTK PSYFQAADDS DTESEPEIED NQGLYAQDPE AEQVEGFIQ GPLDDYADEE VDVVFTSDWK PPELESDEHG KTLRLTSPEG LSGEQKSQWL STIKAVVQSA KYWNLAECTF E ASGEGVIM ...String: MDNLTKVREY LKSYSRLDQA VGEIDEIEAQ RAEKSNYELF QEDGVEEHTK PSYFQAADDS DTESEPEIED NQGLYAQDPE AEQVEGFIQ GPLDDYADEE VDVVFTSDWK PPELESDEHG KTLRLTSPEG LSGEQKSQWL STIKAVVQSA KYWNLAECTF E ASGEGVIM KERQITPDVY KVTPVMNTHP SQSEAVSDVW SLSKTSMTFQ PKKASLQPLT ISLDELFSSR GEFISVGGDG RM SHKEAIL LGLRYKKLYN QARVKYSL UniProtKB: Phosphoprotein |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 3 / Number real images: 3307 / Average exposure time: 5.0 sec. / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 35-2109 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 70.6 / Target criteria: CC + restraints |
| Output model |  PDB-6u1x: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)