[English] 日本語
 Yorodumi
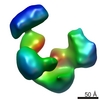
Yorodumi- PDB-1eg0: FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1eg0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RIBOSOME / 70S RIBOSOME / LOW RESOLUTION MODEL | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of translation / large ribosomal subunit / ribosomal small subunit biogenesis / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding ...regulation of translation / large ribosomal subunit / ribosomal small subunit biogenesis / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
| Model details | REFINEMENT DONE BY PROGRAMS O AND SPIDER. ALL COMPONENTS WERE FIT INTO MAP AS RIGID BODIES USING ...REFINEMENT DONE BY PROGRAMS O AND SPIDER. ALL COMPONENTS WERE FIT INTO MAP AS RIGID BODIES USING COORDINATES FROM THE REFERRED SOURCES EXCEPT FOR L1, FOR WHICH ABOUT 15-DEGREE ROTATION WAS INTRODUCED BETWEEN THE TWO DOMAINS; FITTING OF PROTEIN S17 IS RELATIVELY UNCERTAIN; CONFORMATIONAL CHANGES OF ANTICODON AND ACCEPTOR REGIONS OF TRNAFMET UPON BINDING TO THE RIBOSOME WERE NOT MODELED. | |||||||||
 Authors Authors | Gabashvili, I.S. / Agrawal, R.K. / Spahn, C.M.T. / Grassucci, R.A. / Svergun, D.I. / Frank, J. / Penczek, P. | |||||||||
 Citation Citation |  Journal: Cell / Year: 2000 Journal: Cell / Year: 2000Title: Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Authors: I S Gabashvili / R K Agrawal / C M Spahn / R A Grassucci / D I Svergun / J Frank / P Penczek /  Abstract: Over 73,000 projections of the E. coli ribosome bound with formyl-methionyl initiator tRNAf(Met) were used to obtain an 11.5 A cryo-electron microscopy map of the complex. This map allows ...Over 73,000 projections of the E. coli ribosome bound with formyl-methionyl initiator tRNAf(Met) were used to obtain an 11.5 A cryo-electron microscopy map of the complex. This map allows identification of RNA helices, peripheral proteins, and intersubunit bridges. Comparison of double-stranded RNA regions and positions of proteins identified in both cryo-EM and X-ray maps indicates good overall agreement but points to rearrangements of ribosomal components required for the subunit association. Fitting of known components of the 50S stalk base region into the map defines the architecture of the GTPase-associated center and reveals a major change in the orientation of the alpha-sarcin-ricin loop. Analysis of the bridging connections between the subunits provides insight into the dynamic signaling mechanism between the ribosomal subunits. #1:  Journal: Embo J. / Year: 1998 Journal: Embo J. / Year: 1998Title: The Crystal Structure of Ribosomal Protein S4 Reveals a Two-Domain Molecule with an Extensive RNA-Binding Surface: One Domain Shows Structural Homology to the Ets DNA-Binding Motif Authors: Davies, C. / Gerstner, R.B. / Draper, D.E. / Ramakrishnan, V. / White, S.W. #2:  Journal: Nature / Year: 1992 Journal: Nature / Year: 1992Title: The Structure of Ribosomal Protein S5 Reveals Sites of Interaction with 16S Rrna Authors: Ramakrishnan, V. / White, S.W. #3:  Journal: Embo J. / Year: 1994 Journal: Embo J. / Year: 1994Title: Crystal Structure of the Ribosomal Protein S6 from Thermus Thermophilus Authors: Lindahl, M. / Svensson, L.A. / Liljas, A. / Sedelnikova, I.A. / Eliseikina, I.A. / Fomenkova, N.P. / Nevskaya, N. / Nikonov, S.V. / Garber, M.B. / Muranova, T.A. / Rykonova, A.I. / Amons, R. #4:  Journal: Structure / Year: 1997 Journal: Structure / Year: 1997Title: The Structure of Ribosomal Protein S7 at 1.9 A Resolution Reveals a Beta- Hairpin Motif that Binds Double-Stranded Nucleic Acids Authors: Wimberly, B.T. / White, S.W. / Ramakrishnan, V. #5:  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Crystal Structure of Ribosomal Protein S8 from Thermus Thermophilus Reveals a High Degree of Structural Conservation of a Specific RNA Binding Site Authors: Nevskaya, N. / Tischenko, S. / Nikulin, A. / Al-Karadaghi, S. / Liljas, A. / Ehresmann, B. / Ehresmann, C. / Garber, M. / Nikonov, S. #6:  Journal: Structure / Year: 1998 Journal: Structure / Year: 1998Title: Conformational Variability of the N-Terminal Helix in the Structure of Ribosomal Protein S15 Authors: Clemons Jr., W.M. / Davies, C.R. / White, S.W. / Ramakrishnan, V. #7:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Solution Structure of Prokaryotic Ribosomal Protein S17 by High-Resolution NMR Spectroscopy Authors: Jaishree, T. / Ramakrishnan, V. / White, S.W. #8:  Journal: Nature / Year: 1999 Journal: Nature / Year: 1999Title: Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution Authors: Clemons Jr., W.M. / May, J.L.C. / Wimberly, B.T. / Mccutcheon, J.P. / Capel, M.S. / Ramakrishnan, V. #9:  Journal: Embo J. / Year: 1993 Journal: Embo J. / Year: 1993Title: Ribosomal Protein L6: Structural Evidence of Gene Duplication from a Primitive RNA-Binding Protein Authors: Golden, B.L. / Davies, C. / Ramakrishnan, V. / White, S.W. #10:  Journal: Cell(Cambridge,Mass.) / Year: 1999 Journal: Cell(Cambridge,Mass.) / Year: 1999Title: A Detailed View of a Ribosomal Active Site: The Structure of the L11-RNA Complex Authors: Wimberly, B.T. / Guymon, R. / Mccutcheon, J.P. / Ramakrishnan, S.W. / White, V. #11:  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: The Two Faces of the Escherichia Coli 23 S Rrna Sarcin/Ricin Domain: The Structure at 1.11 A Resolution Authors: Correll, C.C. / Wool, I.G. / Munishkin, A. #12:  Journal: Embo J. / Year: 1996 Journal: Embo J. / Year: 1996Title: Crystal Structure of the RNA Binding Ribosomal Prot L1 from Thermus Thermophilus Authors: Nikonov, S. / Nevskaya, N. / Eliseikina, I. / Briand, C. / Al-Karadaghi, S. / Svensson, A. / Liljas, A. / Aebarson, A. #13:  Journal: Embo J. / Year: 1998 Journal: Embo J. / Year: 1998Title: Crystal Structure of Methionyl-Trnafmet Transformylase Complexed with the Initiator Formyl-Methionyl-Trnafmet Authors: Schmitt, E. / Panvert, M. / Blanquet, S. / Mechulam, Y. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1eg0.cif.gz 1eg0.cif.gz | 81.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1eg0.ent.gz pdb1eg0.ent.gz | 42.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1eg0.json.gz 1eg0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eg/1eg0 https://data.pdbj.org/pub/pdb/validation_reports/eg/1eg0 ftp://data.pdbj.org/pub/pdb/validation_reports/eg/1eg0 ftp://data.pdbj.org/pub/pdb/validation_reports/eg/1eg0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1003MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-RNA chain , 4 types, 4 molecules ILMO
| #1: RNA chain | Mass: 6426.170 Da / Num. of mol.: 1 / Fragment: RESIDUES 673-713 / Source method: isolated from a natural source Details: MODELED AS ANALOGOUS FRAGMENT OF T. THERMOPHILUS TAKEN FROM PDB ENTRY 1QD7 Source: (natural)  |
|---|---|
| #2: RNA chain | Mass: 18347.938 Da / Num. of mol.: 1 / Fragment: RESIDUES 1051-1108 / Source method: isolated from a natural source Details: T. MARITIMA RNA SEQUENCE AND MODEL TAKEN FROM PDB ENTRY 1MMS Source: (natural)  |
| #3: RNA chain | Mass: 8438.090 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: E.COLI RNA SEQUENCE AND MODEL TAKEN FROM PDB ENTRY 480D Source: (natural)  |
| #4: RNA chain | Mass: 24526.750 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: E.COLI FMET-TRNA SEQUENCE AND MODEL TAKEN FROM PDB ENTRY 2FMT Source: (natural)  |
-Protein , 8 types, 8 molecules ABCDEFGH
| #5: Protein | Mass: 18624.359 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF B. STEAROTHERMOPHILUS TAKEN FROM PDB ENTRY 1C06 Source: (natural)  |
|---|---|
| #6: Protein | Mass: 15686.305 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF B. STEAROTHERMOPHILUS TAKEN FROM PDB ENTRY 1PKP Source: (natural)  |
| #7: Protein | Mass: 11619.337 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T.THERMOPHILUS TAKEN FROM PDB ENTRY 1RIS Source: (natural)  |
| #8: Protein | Mass: 16758.543 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T. THERMOPHILUS TAKEN FROM PDB ENTRY 1RSS Source: (natural)  |
| #9: Protein | Mass: 15868.570 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T. THERMOPHILUS TAKEN FROM PDB ENTRY 1AN7 Source: (natural)  |
| #10: Protein | Mass: 10578.407 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF B. STEAROTHERMOPHILUS TAKEN FROM PDB ENTRY 1A32 Source: (natural)  |
| #11: Protein | Mass: 10569.341 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF B. STEAROTHERMOPHILUS TAKEN FROM PDB ENTRIES 1RIP AND 1QD7 Source: (natural)  |
| #12: Protein | Mass: 8528.504 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T. THERMOPHILUS TAKEN FROM PDB ENTRY 1QD7 Source: (natural)  |
-PROTEIN (RIBOSOMAL PROTEIN ... , 3 types, 3 molecules NJK
| #13: Protein | Mass: 24867.699 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T. THERMOPHILUS TAKEN FROM PDB ENTRY 1AD2 Source: (natural)  |
|---|---|
| #14: Protein | Mass: 18601.459 Da / Num. of mol.: 1 / Mutation: B. STEAROTHERMOPHILUS SEQUENCE AND MODEL / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T. STEAROTHERMOPHILUS TAKEN FROM PDB ENTRY 1RL6 Source: (natural)  |
| #15: Protein | Mass: 14996.835 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: MODELED BY ANALOGOUS PROTEIN OF T. MARITIMA TAKEN FROM PDB ENTRY 1MMS Source: (natural)  |
-Details
| Sequence details | ONLY COORDINATE |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: E.COLI 70S RIBOSOME / Type: RIBOSOME |
|---|---|
| Buffer solution | pH: 7.6 / Details: 20 mM Hepes, 6 mM Mg(CH3COO)2 150 mM NH4Cl, 4 mM |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Instrument: HOMEMADE PLUNGER / Cryogen name: ETHANE |
| Crystal grow | *PLUS Method: other / Details: Electron Microscopy |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: FEI/PHILIPS CM200FEG/ST / Date: Jan 1, 1997 |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 50000 X / Nominal defocus max: 4340 nm / Nominal defocus min: 730 nm |
| Image recording | Electron dose: 10 e/Å2 / Film or detector model: KODAK SO-163 FILM |
| Reflection | Highest resolution: 11.5 Å |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||
| 3D reconstruction | Resolution: 11.5 Å / Num. of particles: 73523 / Symmetry type: POINT | ||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: RECIPROCAL / Target criteria: VECTOR R-FACTOR Details: REFINEMENT PROTOCOL--RIGID BODY REFINEMENT DETAILS--ALL COMPONENTS WERE FIT INTO MAP AS RIGID BODIES USING COORDINATES FROM THE REFERRED SOURCES EXCEPT FOR L1, FOR WHICH ABOUT 15-DEGREE ...Details: REFINEMENT PROTOCOL--RIGID BODY REFINEMENT DETAILS--ALL COMPONENTS WERE FIT INTO MAP AS RIGID BODIES USING COORDINATES FROM THE REFERRED SOURCES EXCEPT FOR L1, FOR WHICH ABOUT 15-DEGREE ROTATION WAS INTRODUCED BETWEEN THE TWO DOMAINS; FITTING OF PROTEIN S17 IS RELATIVELY UNCERTAIN; CONFORMATIONAL CHANGES OF ANTICODON AND ACCEPTOR REGIONS OF TRNAFMET UPON BINDING TO THE RIBOSOME WERE NOT MODELED. | ||||||||||||
| Refinement | Highest resolution: 11.5 Å Details: ALL COMPONENTS WERE FIT INTO MAP AS RIGID BODIES USING COORDINATES FROM THE REFERRED SOURCES EXCEPT FOR L1, FOR WHICH ABOUT 15-DEGREE ROTATION WAS INTRODUCED BETWEEN THE TWO DOMAINS; FITTING ...Details: ALL COMPONENTS WERE FIT INTO MAP AS RIGID BODIES USING COORDINATES FROM THE REFERRED SOURCES EXCEPT FOR L1, FOR WHICH ABOUT 15-DEGREE ROTATION WAS INTRODUCED BETWEEN THE TWO DOMAINS; FITTING OF PROTEIN S17 IS RELATIVELY UNCERTAIN; CONFORMATIONAL CHANGES OF ANTICODON AND ACCEPTOR REGIONS OF TRNAFMET UPON BINDING TO THE RIBOSOME WERE NOT MODELED. | ||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 11.5 Å
|
 Movie
Movie Controller
Controller











 PDBj
PDBj





























