[English] 日本語
 Yorodumi
Yorodumi- PDB-1elv: CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1elv | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
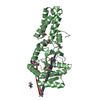
| Title | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S PROTEASE | |||||||||
 Components Components | COMPLEMENT C1S COMPONENT | |||||||||
 Keywords Keywords | HYDROLASE / TRYPSIN-LIKE SERIN PROTEASE / CCP (OR SUSHI OR SCR)MODULE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcomplement subcomponent C_overbar_1s_ / Classical antibody-mediated complement activation / Initial triggering of complement / complement activation, classical pathway / Regulation of Complement cascade / blood microparticle / serine-type endopeptidase activity / innate immune response / calcium ion binding / proteolysis ...complement subcomponent C_overbar_1s_ / Classical antibody-mediated complement activation / Initial triggering of complement / complement activation, classical pathway / Regulation of Complement cascade / blood microparticle / serine-type endopeptidase activity / innate immune response / calcium ion binding / proteolysis / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.7 Å SYNCHROTRON / Resolution: 1.7 Å | |||||||||
 Authors Authors | Gaboriaud, C. / Rossi, V. / Bally, I. / Arlaud, G. / Fontecilla-Camps, J.-C. | |||||||||
 Citation Citation |  Journal: EMBO J. / Year: 2000 Journal: EMBO J. / Year: 2000Title: Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle. Authors: Gaboriaud, C. / Rossi, V. / Bally, I. / Arlaud, G.J. / Fontecilla-Camps, J.C. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1elv.cif.gz 1elv.cif.gz | 84.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1elv.ent.gz pdb1elv.ent.gz | 61.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1elv.json.gz 1elv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1elv_validation.pdf.gz 1elv_validation.pdf.gz | 473.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1elv_full_validation.pdf.gz 1elv_full_validation.pdf.gz | 483.7 KB | Display | |
| Data in XML |  1elv_validation.xml.gz 1elv_validation.xml.gz | 10.3 KB | Display | |
| Data in CIF |  1elv_validation.cif.gz 1elv_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/el/1elv https://data.pdbj.org/pub/pdb/validation_reports/el/1elv ftp://data.pdbj.org/pub/pdb/validation_reports/el/1elv ftp://data.pdbj.org/pub/pdb/validation_reports/el/1elv | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 36687.473 Da / Num. of mol.: 1 Fragment: CCP2-SP CATALYTIC FRAGMENT: ASP363-ASP-673 SEGMENT PRECEDED BY AN ASP-LEU SEQUENCE ADDED AT THE N-TERMINAL END Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Description: BAC-TO-BAC EXPRESSION SYSTEM / Plasmid: PNT-BAC/CCP2-AP-SP / Cell line (production host): Sf21 / Production host: Homo sapiens (human) / Description: BAC-TO-BAC EXPRESSION SYSTEM / Plasmid: PNT-BAC/CCP2-AP-SP / Cell line (production host): Sf21 / Production host:  References: UniProt: P09871, complement subcomponent C_overbar_1s_ | ||||||
|---|---|---|---|---|---|---|---|
| #2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta- ...2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source | ||||||
| #3: Chemical | | #4: Chemical | ChemComp-NES / | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.55 Å3/Da / Density % sol: 51.8 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.4 Details: PEG 4K 34%, AMMONIUM SULFATE 100mM, pH 7.4, VAPOR DIFFUSION, HANGING DROP, temperature 277K | ||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.93 / Beamline: ID14-4 / Wavelength: 0.93 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Sep 24, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.93 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→15 Å / Num. all: 39489 / Num. obs: 115710 / % possible obs: 93 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 2.9 % / Biso Wilson estimate: 21.7 Å2 / Rsym value: 0.037 / Net I/σ(I): 16.9 |
| Reflection shell | Resolution: 1.7→1.75 Å / Redundancy: 1.5 % / Mean I/σ(I) obs: 3.4 / Rsym value: 0.247 / % possible all: 75.8 |
| Reflection | *PLUS Num. obs: 39489 / Num. measured all: 115710 / Rmerge(I) obs: 0.037 |
| Reflection shell | *PLUS % possible obs: 75.8 % / Rmerge(I) obs: 0.247 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.7→15 Å / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Lamzin (arp.dic) Details: LIKELY BECAUSE OF RADIATION DAMAGES, THE INTERCHAIN DISULFIDE BRIDGE (A410-A534) IS NOT FORMED, SEVERAL CYSTEINES ARE OBSERVED WITH DUAL CONFORMATION (A371, A410, A580, A613) AND DENSITY IS ...Details: LIKELY BECAUSE OF RADIATION DAMAGES, THE INTERCHAIN DISULFIDE BRIDGE (A410-A534) IS NOT FORMED, SEVERAL CYSTEINES ARE OBSERVED WITH DUAL CONFORMATION (A371, A410, A580, A613) AND DENSITY IS LACKING AT THE END OF SEVERAL GLU (A351, A359, A372, A373, A385, A397, A402, A506), ASP (A357, A541), LYS (A629) AND MET (A545) RESIDUES. (THIS IS BRIEFLY DISCUSSED IN THE ABOVE REFERENCE).
| |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.7 Å2 | |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→15 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.7 Å / Lowest resolution: 15 Å / σ(F): 0 / Rfactor obs: 0.185 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 26.7 Å2 | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller









 PDBj
PDBj