+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bkc | ||||||
|---|---|---|---|---|---|---|---|
| Title | CATALYTIC DOMAIN OF TNF-ALPHA CONVERTING ENZYME (TACE) | ||||||
 Components Components | (TUMOR NECROSIS FACTOR-ALPHA-CONVERTING ...) x 3 | ||||||
 Keywords Keywords | ZN-ENDOPEPTIDASE / HYDROLASE / TNF-ALPHA | ||||||
| Function / homology |  Function and homology information Function and homology informationMetallo-peptidase family M12 / Domain of unknown function DUF3850 / Domain of unknown function (DUF3850) / ASCH / ASCH domain / Collagenase (Catalytic Domain) / Collagenase (Catalytic Domain) / PUA-like superfamily / 3-Layer(aba) Sandwich / Alpha Beta Similarity search - Domain/homology | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / MULTIWAVELENGTH ANOMALOUS DISPERSION / Resolution: 2 Å SYNCHROTRON / MULTIWAVELENGTH ANOMALOUS DISPERSION / Resolution: 2 Å | ||||||
 Authors Authors | Maskos, K. / Fernandez-Catalan, C. / Bode, W. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 1998 Journal: Proc.Natl.Acad.Sci.USA / Year: 1998Title: Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Authors: Maskos, K. / Fernandez-Catalan, C. / Huber, R. / Bourenkov, G.P. / Bartunik, H. / Ellestad, G.A. / Reddy, P. / Wolfson, M.F. / Rauch, C.T. / Castner, B.J. / Davis, R. / Clarke, H.R. / ...Authors: Maskos, K. / Fernandez-Catalan, C. / Huber, R. / Bourenkov, G.P. / Bartunik, H. / Ellestad, G.A. / Reddy, P. / Wolfson, M.F. / Rauch, C.T. / Castner, B.J. / Davis, R. / Clarke, H.R. / Petersen, M. / Fitzner, J.N. / Cerretti, D.P. / March, C.J. / Paxton, R.J. / Black, R.A. / Bode, W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bkc.cif.gz 1bkc.cif.gz | 257 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bkc.ent.gz pdb1bkc.ent.gz | 202 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bkc.json.gz 1bkc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bk/1bkc https://data.pdbj.org/pub/pdb/validation_reports/bk/1bkc ftp://data.pdbj.org/pub/pdb/validation_reports/bk/1bkc ftp://data.pdbj.org/pub/pdb/validation_reports/bk/1bkc | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 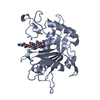
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
-TUMOR NECROSIS FACTOR-ALPHA-CONVERTING ... , 3 types, 4 molecules ACEI
| #1: Protein | Mass: 28954.457 Da / Num. of mol.: 2 / Mutation: S266A, N452Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / References: UniProt: P78536 Homo sapiens (human) / References: UniProt: P78536#2: Protein | | Mass: 28970.480 Da / Num. of mol.: 1 / Mutation: S266A, N452Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / References: UniProt: P78536 Homo sapiens (human) / References: UniProt: P78536#3: Protein | | Mass: 28968.484 Da / Num. of mol.: 1 / Mutation: S266A, N452Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / References: UniProt: P78536 Homo sapiens (human) / References: UniProt: P78536 |
|---|
-Non-polymers , 3 types, 1628 molecules 



| #4: Chemical | ChemComp-ZN / #5: Chemical | ChemComp-INN / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.59 Å3/Da / Density % sol: 52.6 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 5.4 / Details: pH 5.4 | ||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, sitting drop | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: MPG/DESY, HAMBURG SYNCHROTRON / Site: MPG/DESY, HAMBURG  / Beamline: BW6 / Wavelength: 1.2776 / Beamline: BW6 / Wavelength: 1.2776 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Aug 1, 1997 / Details: MIRRORS |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.2776 Å / Relative weight: 1 |
| Reflection | Resolution: 2→20 Å / Num. obs: 77653 / % possible obs: 96.9 % / Observed criterion σ(I): 2 / Redundancy: 5 % / Rmerge(I) obs: 0.031 |
| Reflection shell | Resolution: 2→2 Å |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: MULTIWAVELENGTH ANOMALOUS DISPERSION Resolution: 2→12 Å /
| ||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→12 Å
|
 Movie
Movie Controller
Controller













 PDBj
PDBj


