[English] 日本語
 Yorodumi
Yorodumi- EMDB-18207: Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc3... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide, Glacios map | |||||||||
 Map data Map data | DeepEMhancer map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ubiquitin / Ubiquitin priming / mono-ubiquitination / CUL2 / UBE2R2 / CDC34 / ELOB/C / FEM1C / monoubiquitination cullin / CRL / cullin-RING ligase / E3 / UBE2R / LIGASE / SIL1 | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
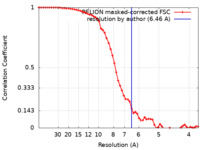
| Method | single particle reconstruction / cryo EM / Resolution: 6.46 Å | |||||||||
 Authors Authors | Liwocha J / Prabu JR / Kleiger G / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting. Authors: Jerry Li / Nicholas Purser / Joanna Liwocha / Daniel C Scott / Holly A Byers / Barbara Steigenberger / Spencer Hill / Ishita Tripathi-Giesgen / Trent Hinkle / Fynn M Hansen / J Rajan Prabu / ...Authors: Jerry Li / Nicholas Purser / Joanna Liwocha / Daniel C Scott / Holly A Byers / Barbara Steigenberger / Spencer Hill / Ishita Tripathi-Giesgen / Trent Hinkle / Fynn M Hansen / J Rajan Prabu / Senthil K Radhakrishnan / Donald S Kirkpatrick / Kurt M Reichermeier / Brenda A Schulman / Gary Kleiger /   Abstract: Cullin-RING ligases (CRLs) ubiquitylate specific substrates selected from other cellular proteins. Substrate discrimination and ubiquitin transferase activity were thought to be strictly separated. ...Cullin-RING ligases (CRLs) ubiquitylate specific substrates selected from other cellular proteins. Substrate discrimination and ubiquitin transferase activity were thought to be strictly separated. Substrates are recognized by substrate receptors, such as Fbox or BCbox proteins. Meanwhile, CRLs employ assorted ubiquitin-carrying enzymes (UCEs, which are a collection of E2 and ARIH-family E3s) specialized for either initial substrate ubiquitylation (priming) or forging poly-ubiquitin chains. We discovered specific human CRL-UCE pairings governing substrate priming. The results reveal pairing of CUL2-based CRLs and UBE2R-family UCEs in cells, essential for efficient PROTAC-induced neo-substrate degradation. Despite UBE2R2's intrinsic programming to catalyze poly-ubiquitylation, CUL2 employs this UCE for geometrically precise PROTAC-dependent ubiquitylation of a neo-substrate and for rapid priming of substrates recruited to diverse receptors. Cryo-EM structures illuminate how CUL2-based CRLs engage UBE2R2 to activate substrate ubiquitylation. Thus, pairing with a specific UCE overcomes E2 catalytic limitations to drive substrate ubiquitylation and targeted protein degradation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18207.map.gz emd_18207.map.gz | 16.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18207-v30.xml emd-18207-v30.xml emd-18207.xml emd-18207.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18207_fsc.xml emd_18207_fsc.xml | 6.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_18207.png emd_18207.png | 42.2 KB | ||
| Masks |  emd_18207_msk_1.map emd_18207_msk_1.map | 18.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18207.cif.gz emd-18207.cif.gz | 4.2 KB | ||
| Others |  emd_18207_additional_1.map.gz emd_18207_additional_1.map.gz emd_18207_half_map_1.map.gz emd_18207_half_map_1.map.gz emd_18207_half_map_2.map.gz emd_18207_half_map_2.map.gz | 1.9 MB 13.8 MB 13.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18207 http://ftp.pdbj.org/pub/emdb/structures/EMD-18207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18207 | HTTPS FTP |
-Validation report
| Summary document |  emd_18207_validation.pdf.gz emd_18207_validation.pdf.gz | 609 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18207_full_validation.pdf.gz emd_18207_full_validation.pdf.gz | 608.5 KB | Display | |
| Data in XML |  emd_18207_validation.xml.gz emd_18207_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_18207_validation.cif.gz emd_18207_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18207 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18207 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18207 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18207 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18207.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18207.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.885 Å | ||||||||||||||||||||||||||||||||||||
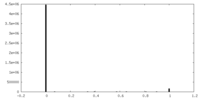
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18207_msk_1.map emd_18207_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
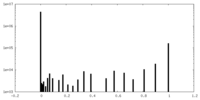
| Density Histograms |
-Additional map: Relion postprocessed map
| File | emd_18207_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion postprocessed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18207_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
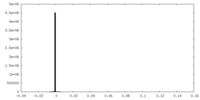
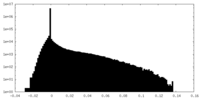
| Density Histograms |
-Half map: #2
| File | emd_18207_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc3...
| Entire | Name: Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide |
|---|---|
| Components |
|
-Supramolecule #1: Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc3...
| Supramolecule | Name: Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 210 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 22000 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)