[English] 日本語
 Yorodumi
Yorodumi- EMDB-16836: Structure of the Pol II-SPT6-Elongin complex (structure 1, global map) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Pol II-SPT6-Elongin complex (structure 1, global map) | ||||||||||||
 Map data Map data | Global map for Pol II-SPT6-Elongin structure | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Transcription elongation / Elongin / RNA polymerase II / TRANSCRIPTION | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | ||||||||||||
 Authors Authors | Chen Y / Kokic G / Dienemann C / Dybkov O / Urlaub H / Cramer P | ||||||||||||
| Funding support | European Union,  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure of the transcribing RNA polymerase II-Elongin complex. Authors: Ying Chen / Goran Kokic / Christian Dienemann / Olexandr Dybkov / Henning Urlaub / Patrick Cramer /   Abstract: Elongin is a heterotrimeric elongation factor for RNA polymerase (Pol) II transcription that is conserved among metazoa. Here, we report three cryo-EM structures of human Elongin bound to ...Elongin is a heterotrimeric elongation factor for RNA polymerase (Pol) II transcription that is conserved among metazoa. Here, we report three cryo-EM structures of human Elongin bound to transcribing Pol II. The structures show that Elongin subunit ELOA binds the RPB2 side of Pol II and anchors the ELOB-ELOC subunit heterodimer. ELOA contains a 'latch' that binds between the end of the Pol II bridge helix and funnel helices, thereby inducing a conformational change near the polymerase active center. The latch is required for the elongation-stimulatory activity of Elongin, but not for Pol II binding, indicating that Elongin functions by allosterically regulating the conformational mobility of the polymerase active center. Elongin binding to Pol II is incompatible with association of the super elongation complex, PAF1 complex and RTF1, which also contain an elongation-stimulatory latch element. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16836.map.gz emd_16836.map.gz | 301.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16836-v30.xml emd-16836-v30.xml emd-16836.xml emd-16836.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16836.png emd_16836.png | 95.2 KB | ||
| Masks |  emd_16836_msk_1.map emd_16836_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16836.cif.gz emd-16836.cif.gz | 4.3 KB | ||
| Others |  emd_16836_half_map_1.map.gz emd_16836_half_map_1.map.gz emd_16836_half_map_2.map.gz emd_16836_half_map_2.map.gz | 261.7 MB 261.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16836 http://ftp.pdbj.org/pub/emdb/structures/EMD-16836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16836 | HTTPS FTP |
-Validation report
| Summary document |  emd_16836_validation.pdf.gz emd_16836_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16836_full_validation.pdf.gz emd_16836_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16836_validation.xml.gz emd_16836_validation.xml.gz | 16.7 KB | Display | |
| Data in CIF |  emd_16836_validation.cif.gz emd_16836_validation.cif.gz | 20 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16836 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16836 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16836 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16836 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16836.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16836.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global map for Pol II-SPT6-Elongin structure | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
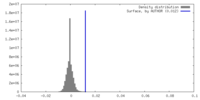
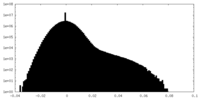
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16836_msk_1.map emd_16836_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
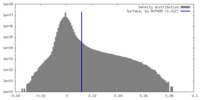
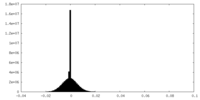
| Density Histograms |
-Half map: half map 1 for generating the global map
| File | emd_16836_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 for generating the global map | ||||||||||||
| Projections & Slices |
| ||||||||||||
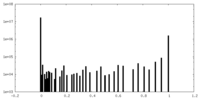
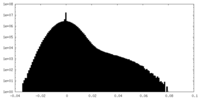
| Density Histograms |
-Half map: half map 2 for generating the global map
| File | emd_16836_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 for generating the global map | ||||||||||||
| Projections & Slices |
| ||||||||||||
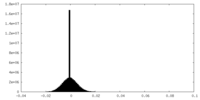
| Density Histograms |
- Sample components
Sample components
-Entire : The Pol II-SPT6-Elongin transcription elongation complex
| Entire | Name: The Pol II-SPT6-Elongin transcription elongation complex |
|---|---|
| Components |
|
-Supramolecule #1: The Pol II-SPT6-Elongin transcription elongation complex
| Supramolecule | Name: The Pol II-SPT6-Elongin transcription elongation complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#19 Details: The map is generated from a sub-class of a Cryo-EM dataset for the Pol II-SPT6-Elongin complex. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.1 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.09 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 7.5 µm / Nominal defocus min: 0.35000000000000003 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.05 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 72087 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)