[English] 日本語
 Yorodumi
Yorodumi- EMDB-12861: Apo-structure of Lassa virus L protein (well-resolved alpha ribbo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12861 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo-structure of Lassa virus L protein (well-resolved alpha ribbon) [APO-RIBBON] | |||||||||
 Map data Map data | Apo-structure of Lassa virus L protein (well-resolved alpha ribbon) [APO-RIBBON] | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lassa virus RNA-dependent RNA polymerase viral RNA / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA replication / cap snatching / virion component / host cell cytoplasm / Hydrolases; Acting on ester bonds / hydrolase activity / RNA-directed RNA polymerase / RNA-dependent RNA polymerase activity / nucleotide binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lassa mammarenavirus Lassa mammarenavirus | |||||||||
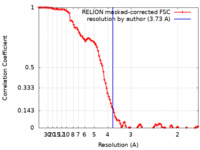
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Kouba T / Vogel D | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Authors: Tomas Kouba / Dominik Vogel / Sigurdur R Thorkelsson / Emmanuelle R J Quemin / Harry M Williams / Morlin Milewski / Carola Busch / Stephan Günther / Kay Grünewald / Maria Rosenthal / Stephen Cusack /   Abstract: Lassa virus is endemic in West Africa and can cause severe hemorrhagic fever. The viral L protein transcribes and replicates the RNA genome via its RNA-dependent RNA polymerase activity. Here, we ...Lassa virus is endemic in West Africa and can cause severe hemorrhagic fever. The viral L protein transcribes and replicates the RNA genome via its RNA-dependent RNA polymerase activity. Here, we present nine cryo-EM structures of the L protein in the apo-, promoter-bound pre-initiation and active RNA synthesis states. We characterize distinct binding pockets for the conserved 3' and 5' promoter RNAs and show how full-promoter binding induces a distinct pre-initiation conformation. In the apo- and early elongation states, the endonuclease is inhibited by two distinct L protein peptides, whereas in the pre-initiation state it is uninhibited. In the early elongation state, a template-product duplex is bound in the active site cavity together with an incoming non-hydrolysable nucleotide and the full C-terminal region of the L protein, including the putative cap-binding domain, is well-ordered. These data advance our mechanistic understanding of how this flexible and multifunctional molecular machine is activated. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12861.map.gz emd_12861.map.gz | 51 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12861-v30.xml emd-12861-v30.xml emd-12861.xml emd-12861.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12861_fsc.xml emd_12861_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12861.png emd_12861.png | 106.4 KB | ||
| Filedesc metadata |  emd-12861.cif.gz emd-12861.cif.gz | 6.8 KB | ||
| Others |  emd_12861_additional_1.map.gz emd_12861_additional_1.map.gz emd_12861_half_map_1.map.gz emd_12861_half_map_1.map.gz emd_12861_half_map_2.map.gz emd_12861_half_map_2.map.gz | 96.2 MB 80.9 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12861 http://ftp.pdbj.org/pub/emdb/structures/EMD-12861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12861 | HTTPS FTP |
-Validation report
| Summary document |  emd_12861_validation.pdf.gz emd_12861_validation.pdf.gz | 772.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12861_full_validation.pdf.gz emd_12861_full_validation.pdf.gz | 772.3 KB | Display | |
| Data in XML |  emd_12861_validation.xml.gz emd_12861_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_12861_validation.cif.gz emd_12861_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12861 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12861 | HTTPS FTP |
-Related structure data
| Related structure data |  7oe7MC  7ochC  7oe3C  7oeaC  7oebC  7ojjC  7ojkC  7ojlC  7ojnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12861.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12861.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apo-structure of Lassa virus L protein (well-resolved alpha ribbon) [APO-RIBBON] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
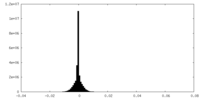
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_12861_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
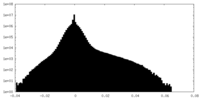
| Density Histograms |
-Half map: #2
| File | emd_12861_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
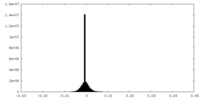
| Density Histograms |
-Half map: #1
| File | emd_12861_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
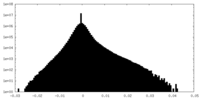
| Density Histograms |
- Sample components
Sample components
-Entire : RNA-directed RNA polymerase L Lassa mammarenavirus
| Entire | Name: RNA-directed RNA polymerase L Lassa mammarenavirus |
|---|---|
| Components |
|
-Supramolecule #1: RNA-directed RNA polymerase L Lassa mammarenavirus
| Supramolecule | Name: RNA-directed RNA polymerase L Lassa mammarenavirus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 253.346 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 253.656938 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEDDMACVKD LVSKYLADNE RLSRQKLAFL VQTEPRMLLM EGLKLLSLCI EIDSCNANGC EHNSEDKSVE RILHDHGILT PSLCFVVPD GYKLTGNVLI LLECFVRSSP ANFEQKYIED FKKLEQLKED LKSVDINLIP LIDGRTSFYN EQIPDWVNDK L RDTLFSLL ...String: MEDDMACVKD LVSKYLADNE RLSRQKLAFL VQTEPRMLLM EGLKLLSLCI EIDSCNANGC EHNSEDKSVE RILHDHGILT PSLCFVVPD GYKLTGNVLI LLECFVRSSP ANFEQKYIED FKKLEQLKED LKSVDINLIP LIDGRTSFYN EQIPDWVNDK L RDTLFSLL RYAQESNSLF EESEYSRLCE SLSMTSGRLS GVESLNVLLD NRSSHYEEII ASCHQGINNK LTAHEVKLQI EE EYQVFRN RLRKGEITGQ FLKVDKSRLL NDFNNLYVDE VTATKDNIEH LIYQFKRASP ILRFLYANIG EGNGEERHHT IKE CQMQYW RSFLNKVKSL RILNTRRKLL LIFDALILLA SIHDQTRHKC SKGWLGSCFI SVNDRLVSLE STKRDLEKWV GRRQ QSERS NTIQPPDKNQ ILISMFQKTI LKATAALKDV GISVEHYKIN MEVICPDSYD LILNFDVSGV VPTISYQRTE DEKFP FIMG GVELLESTDL ERLSSLSLAL VNSMKTSSTV KLRQNEFGPA RYQVVRCKEA YCQEFLLSGA EFQLIYQKTG ECSKCY AIN DNRVGEICSF YADPKRYFPA IFSAEVLQTT VSTMISWVKD CSELEEQLCN INSLTKMILV LILAHPSKRS QKLLQNL RY FIMAYVSDYH HKDLIDKLRE ELITDVEFLL YRLVRALVNL ILSEDVKSMM TNRFKFILNI SYMCHFITKE TPDRLTDQ I KCFEKFLEPK LEFGHVSINP ADVATEEELD DMVYNAKKFL SKEGCTSIKG PDYKKPGVSK RFLSLLTSSF NNGSLFKES EVKREIKDPL VTSGCATALD LASNKSVVVN KYTDGSRVLN YDFNKLTALA VSQLTEVFSR KGKHLLNKQD YDYKVQQAMS NLVLGPRQN KVGADEADLD EILLDGGASV YFDQLKETVE RIIDQYREPV KPGSNPNGGD QPSVNDLDEV VPNKFYIRLI K GELSNHMV EDFDYDVLPG NFYEEFCDAV YKNNKLKERY FYCGQMSQCP IGELTKAVAT RTYFDQEYFQ CFKSILLIMN AN TLMGRYT HYKSRNLNFK FDMGRLSDDV RISERESNSE ALSKALSLTN CTTAMLKNLC FYSQESPQSY SSTGPDTGRL KFS LSYKEQ VGGNRELYIG DLRTKMFTRL IEDYFEALSL QLSGSCLNNE REFENAILSM KLNVSLAHVS YSMDHSKWGP MMCP FLFLA TLQNLIFLSK DLQADIKGRD YLSTLLTWHM HKMVEIPFNV VSAMMKSFIK AQLGLKKKTT QSITEDFFYS NFQIG VVPS HVSSILDMGQ GILHNTSDFY ALISERFINY AISCICGGTI DAYTSSDDQI SLFDQVLTEL MQRDPEEFKT LIEFHY YMS DQLNKFVSPK SVIGRFVAEF KSRFYVWGDE VPLLTKFVAA ALHNIKCKEP HQLAETIDTI IDQSVANGVP VHLCNLI QK RTLSLLQYAR YPIDPFLLNC ETDVRDWVDG NRSYRIMRQI ERLIPDACGR IRSMLRKLYN KLKTGQLHEE FTTNYLSS E HLSSLSNLCE LLGVEPPSES DLEFSWLNLA AHHPLRMVLR QKIIYSGAVN LDDEKVPTIV KTIQNKLSST FTRGAQKLL SEAINKSAFQ SSIASGFVGL CRTLGSKCVR GPNKESLYIK SIQSLISDIQ GIEPLIDSHG VQYWRVPLNI RDGNEGVISY FRPLLWDYM CISLSTAIEL GAWVLGEPKK VRVLEFFKHN PCDYFPLKPA ASKLLEDRVG LNHIIHSLRR LYPSVFEKHI L PFMSDLAS TKMKWSPRIK FLDLCVALDV NCEALSLVSH IVKWKREEHY IVLSSELRLS HTRTHEPMVE ERVVSTSDAV DN FMRQIYF ESYVRSFVAT TRTLGSFTWF PHKTSVPEGE GLQRLGPFSS FVEKAIHKGI ERPMFKHDLM MGYAWIDFDI EPA RFNHNQ LIASGLVGPR FDSLEDFFDA VESLPPGSAK LSQTVRFRIK SQDASFKESF AIHLDYTGSI NQQTKYLVHE VSAM YSGAV SPCVLSDCWR LVLSGPTFKG KSAWYVDTEI VNEFLTDTNQ LGHVTPVEIV VDMEKLQFTE YDFVLVGPCV EPVPL VVHR GGLWECDKKL ASFTPVVQDQ DLEMFVKEVG DSSLDLLIGA LSAMILDRLK LRMQWSEVDI VSMLKAAMPS NSVKVL NAV LEAVDDWVDF KGYALCYSKS RKKVMVHSSG GKLRLKGRTC EELVKEDEGI EDIE UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)