+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12588 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of unliganded O-GlcNAc transferase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | O-GlcNAc transferase O-linked B-n-acetylglucosamine transferase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein N-acetylglucosaminyltransferase complex / protein O-GlcNAc transferase / regulation of insulin receptor signaling pathway / protein O-acetylglucosaminyltransferase activity / positive regulation of transcription from RNA polymerase II promoter by glucose / acetylglucosaminyltransferase activity / regulation of necroptotic process / regulation of Rac protein signal transduction / negative regulation of stem cell population maintenance / protein O-linked glycosylation ...protein N-acetylglucosaminyltransferase complex / protein O-GlcNAc transferase / regulation of insulin receptor signaling pathway / protein O-acetylglucosaminyltransferase activity / positive regulation of transcription from RNA polymerase II promoter by glucose / acetylglucosaminyltransferase activity / regulation of necroptotic process / regulation of Rac protein signal transduction / negative regulation of stem cell population maintenance / protein O-linked glycosylation / NSL complex / regulation of glycolytic process / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / RIPK1-mediated regulated necrosis / regulation of synapse assembly / regulation of gluconeogenesis / positive regulation of stem cell population maintenance / Formation of WDR5-containing histone-modifying complexes / phosphatidylinositol-3,4,5-trisphosphate binding / positive regulation of proteolysis / mitophagy / hemopoiesis / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / histone acetyltransferase complex / positive regulation of lipid biosynthetic process / negative regulation of protein ubiquitination / positive regulation of TORC1 signaling / negative regulation of cell migration / response to nutrient / cell projection / positive regulation of translation / mitochondrial membrane / cellular response to glucose stimulus / negative regulation of transforming growth factor beta receptor signaling pathway / circadian regulation of gene expression / response to insulin / Regulation of necroptotic cell death / protein processing / chromatin DNA binding / UCH proteinases / chromatin organization / positive regulation of cold-induced thermogenesis / HATs acetylate histones / glutamatergic synapse / apoptotic process / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex / nucleoplasm / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
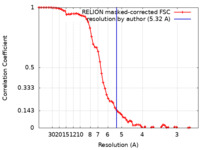
| Method | single particle reconstruction / cryo EM / Resolution: 5.32 Å | |||||||||
 Authors Authors | Meek RW / Blaza JN | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structure provides insights into the dimer arrangement of the O-linked β-N-acetylglucosamine transferase OGT. Authors: Richard W Meek / James N Blaza / Jil A Busmann / Matthew G Alteen / David J Vocadlo / Gideon J Davies /   Abstract: The O-linked β-N-acetylglucosamine modification is a core signalling mechanism, with erroneous patterns leading to cancer and neurodegeneration. Although thousands of proteins are subject to this ...The O-linked β-N-acetylglucosamine modification is a core signalling mechanism, with erroneous patterns leading to cancer and neurodegeneration. Although thousands of proteins are subject to this modification, only a single essential glycosyltransferase catalyses its installation, the O-GlcNAc transferase, OGT. Previous studies have provided truncated structures of OGT through X-ray crystallography, but the full-length protein has never been observed. Here, we report a 5.3 Å cryo-EM model of OGT. We show OGT is a dimer, providing a structural basis for how some X-linked intellectual disability mutations at the interface may contribute to disease. We observe that the catalytic section of OGT abuts a 13.5 tetratricopeptide repeat unit region and find the relative positioning of these sections deviate from the previously proposed, X-ray crystallography-based model. We also note that OGT exhibits considerable heterogeneity in tetratricopeptide repeat units N-terminal to the dimer interface with repercussions for how OGT binds protein ligands and partners. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12588.map.gz emd_12588.map.gz | 25.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12588-v30.xml emd-12588-v30.xml emd-12588.xml emd-12588.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12588_fsc.xml emd_12588_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12588.png emd_12588.png | 110.3 KB | ||
| Filedesc metadata |  emd-12588.cif.gz emd-12588.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12588 http://ftp.pdbj.org/pub/emdb/structures/EMD-12588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12588 | HTTPS FTP |
-Validation report
| Summary document |  emd_12588_validation.pdf.gz emd_12588_validation.pdf.gz | 533.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12588_full_validation.pdf.gz emd_12588_full_validation.pdf.gz | 533.4 KB | Display | |
| Data in XML |  emd_12588_validation.xml.gz emd_12588_validation.xml.gz | 9 KB | Display | |
| Data in CIF |  emd_12588_validation.cif.gz emd_12588_validation.cif.gz | 11.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12588 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12588 | HTTPS FTP |
-Related structure data
| Related structure data |  7ntfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12588.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12588.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.302 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
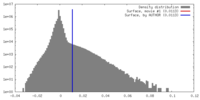
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dimeric assembly of O-GlcNAc transferase
| Entire | Name: Dimeric assembly of O-GlcNAc transferase |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric assembly of O-GlcNAc transferase
| Supramolecule | Name: Dimeric assembly of O-GlcNAc transferase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Isoform 1 of UDP-N-acetylglucosamine--peptide N-acetylglucosaminy...
| Macromolecule | Name: Isoform 1 of UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: protein O-GlcNAc transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120.225773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEFELRR QASSVGNVAD STGLAELAHR EYQAGDFEAA ERHCMQLWRQ EPDNTGVLL LLSSIHFQCR RLDRSAHFST LAIKQNPLLA EAYSNLGNVY KERGQLQEAI EHYRHALRLK PDFIDGYINL A AALVAAGD ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEFELRR QASSVGNVAD STGLAELAHR EYQAGDFEAA ERHCMQLWRQ EPDNTGVLL LLSSIHFQCR RLDRSAHFST LAIKQNPLLA EAYSNLGNVY KERGQLQEAI EHYRHALRLK PDFIDGYINL A AALVAAGD MEGAVQAYVS ALQYNPDLYC VRSDLGNLLK ALGRLEEAKA CYLKAIETQP NFAVAWSNLG CVFNAQGEIW LA IHHFEKA VTLDPNFLDA YINLGNVLKE ARIFDRAVAA YLRALSLSPN HAVVHGNLAC VYYEQGLIDL AIDTYRRAIE LQP HFPDAY CNLANALKEK GSVAEAEDCY NTALRLCPTH ADSLNNLANI KREQGNIEEA VRLYRKALEV FPEFAAAHSN LASV LQQQG KLQEALMHYK EAIRISPTFA DAYSNMGNTL KEMQDVQGAL QCYTRAIQIN PAFADAHSNL ASIHKDSGNI PEAIA SYRT ALKLKPDFPD AYCNLAHCLQ IVCDWTDYDE RMKKLVSIVA DQLEKNRLPS VHPHHSMLYP LSHGFRKAIA ERHGNL CLD KINVLHKPPY EHPKDLKLSD GRLRVGYVSS DFGNHPTSHL MQSIPGMHNP DKFEVFCYAL SPDDGTNFRV KVMAEAN HF IDLSQIPCNG KAADRIHQDG IHILVNMNGY TKGARNELFA LRPAPIQAMW LGYPGTSGAL FMDYIITDQE TSPAEVAE Q YSEKLAYMPH TFFIGDHANM FPHLKKKAVI DFKSNGHIYD NRIVLNGIDL KAFLDSLPDV KIVKMKCPDG GDNADSSNT ALNMPVIPMN TIAEAVIEMI NRGQIQITIN GFSISNGLAT TQINNKAATG EEVPRTIIVT TRSQYGLPED AIVYCNFNQL YKIDPSTLQ MWANILKRVP NSVLWLLRFP AVGEPNIQQY AQNMGLPQNR IIFSPVAPKE EHVRRGQLAD VCLDTPLCNG H TTGMDVLW AGTPMVTMPG ETLASRVAAS QLTCLGCLEL IAKNRQEYED IAVKLGTDLE YLKKVRGKVW KQRISSPLFN TK QYTMELE RLYLQMWEHY AAGNKPDHMI KPVEVTESA UniProtKB: UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25 mM HEPES pH 7.5 150 mM NaCl 1 mM DTT 0.5 % glycerol |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 180 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.038 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 1.5 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)