[English] 日本語
 Yorodumi
Yorodumi- EMDB-12292: In situ structure of the I-band thin filament excluding troponin ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12292 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of the I-band thin filament excluding troponin from mouse psoas muscle | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 10.6 Å | ||||||||||||||||||
 Authors Authors | Wang Z / Grange M / Wagner T / Kho AL / Gautel M / Raunser S | ||||||||||||||||||
| Funding support |  Germany, Germany,  United Kingdom, European Union, 5 items United Kingdom, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: The molecular basis for sarcomere organization in vertebrate skeletal muscle. Authors: Zhexin Wang / Michael Grange / Thorsten Wagner / Ay Lin Kho / Mathias Gautel / Stefan Raunser /   Abstract: Sarcomeres are force-generating and load-bearing devices of muscles. A precise molecular picture of how sarcomeres are built underpins understanding their role in health and disease. Here, we ...Sarcomeres are force-generating and load-bearing devices of muscles. A precise molecular picture of how sarcomeres are built underpins understanding their role in health and disease. Here, we determine the molecular architecture of native vertebrate skeletal sarcomeres by electron cryo-tomography. Our reconstruction reveals molecular details of the three-dimensional organization and interaction of actin and myosin in the A-band, I-band, and Z-disc and demonstrates that α-actinin cross-links antiparallel actin filaments by forming doublets with 6-nm spacing. Structures of myosin, tropomyosin, and actin at ~10 Å further reveal two conformations of the "double-head" myosin, where the flexible orientation of the lever arm and light chains enable myosin not only to interact with the same actin filament, but also to split between two actin filaments. Our results provide unexpected insights into the fundamental organization of vertebrate skeletal muscle and serve as a strong foundation for future investigations of muscle diseases. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12292.map.gz emd_12292.map.gz | 912.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12292-v30.xml emd-12292-v30.xml emd-12292.xml emd-12292.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12292_fsc.xml emd_12292_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12292.png emd_12292.png | 31.6 KB | ||
| Masks |  emd_12292_msk_1.map emd_12292_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Others |  emd_12292_half_map_1.map.gz emd_12292_half_map_1.map.gz emd_12292_half_map_2.map.gz emd_12292_half_map_2.map.gz | 2.8 MB 2.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12292 http://ftp.pdbj.org/pub/emdb/structures/EMD-12292 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12292 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12292 | HTTPS FTP |
-Validation report
| Summary document |  emd_12292_validation.pdf.gz emd_12292_validation.pdf.gz | 355.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12292_full_validation.pdf.gz emd_12292_full_validation.pdf.gz | 354.8 KB | Display | |
| Data in XML |  emd_12292_validation.xml.gz emd_12292_validation.xml.gz | 9.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12292 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12292 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12292 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12292 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12292.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12292.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.51 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
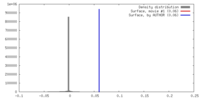
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12292_msk_1.map emd_12292_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
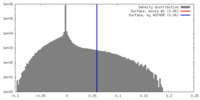
| Density Histograms |
-Half map: #2
| File | emd_12292_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12292_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : I-band thin filament excluding troponin from mouse psoas muscle
| Entire | Name: I-band thin filament excluding troponin from mouse psoas muscle |
|---|---|
| Components |
|
-Supramolecule #1: I-band thin filament excluding troponin from mouse psoas muscle
| Supramolecule | Name: I-band thin filament excluding troponin from mouse psoas muscle type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: actin
| Supramolecule | Name: actin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: tropomyosin
| Supramolecule | Name: tropomyosin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4-#6 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | The sample was myofibrils. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-8 / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 28409 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller




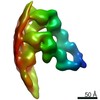









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)