[English] 日本語
 Yorodumi
Yorodumi- EMDB-3128: Structure of a cross-beta amyloid fibril from IGSNVVTWYQQL peptid... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3128 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a cross-beta amyloid fibril from IGSNVVTWYQQL peptide of AL-DIA immunoglobulin light chain by cryo-EM and fiber diffraction | |||||||||
 Map data Map data | Reconstruction morphology 1 of a left-handed amyloid-like fibril of the IGSNVVTWYQQL fragment of an immunoglobulin light chain | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AL amyloidosis / cryo electron microscopy / steric zipper / three dimensional reconstruction | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
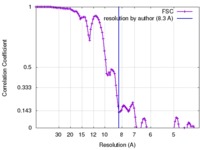
| Method | helical reconstruction / cryo EM / Resolution: 8.3 Å | |||||||||
 Authors Authors | Schmidt A / Annamalai K / Schmidt M / Grigorieff N / Fandrich M | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: Cryo-EM reveals the steric zipper structure of a light chain-derived amyloid fibril. Authors: Andreas Schmidt / Karthikeyan Annamalai / Matthias Schmidt / Nikolaus Grigorieff / Marcus Fändrich /   Abstract: Amyloid fibrils are proteinaceous aggregates associated with diseases in humans and animals. The fibrils are defined by intermolecular interactions between the fibril-forming polypeptide chains, but ...Amyloid fibrils are proteinaceous aggregates associated with diseases in humans and animals. The fibrils are defined by intermolecular interactions between the fibril-forming polypeptide chains, but it has so far remained difficult to reveal the assembly of the peptide subunits in a full-scale fibril. Using electron cryomicroscopy (cryo-EM), we present a reconstruction of a fibril formed from the pathogenic core of an amyloidogenic immunoglobulin (Ig) light chain. The fibril density shows a lattice-like assembly of face-to-face packed peptide dimers that corresponds to the structure of steric zippers in peptide crystals. Interpretation of the density map with a molecular model enabled us to identify the intermolecular interactions between the peptides and rationalize the hierarchical structure of the fibril based on simple chemical principles. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3128.map.gz emd_3128.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3128-v30.xml emd-3128-v30.xml emd-3128.xml emd-3128.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3128_fsc.xml emd_3128_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_3128.png emd_3128.png | 16.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3128 http://ftp.pdbj.org/pub/emdb/structures/EMD-3128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3128 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3128.map.gz / Format: CCP4 / Size: 2.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3128.map.gz / Format: CCP4 / Size: 2.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction morphology 1 of a left-handed amyloid-like fibril of the IGSNVVTWYQQL fragment of an immunoglobulin light chain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain...
| Entire | Name: Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain of Human AL Patient |
|---|---|
| Components |
|
-Supramolecule #1000: Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain...
| Supramolecule | Name: Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain of Human AL Patient type: sample / ID: 1000 / Details: The sample forms long amyloid-like fibrils. / Number unique components: 1 |
|---|
-Macromolecule #1: Immunoglobulin Light Chain
| Macromolecule | Name: Immunoglobulin Light Chain / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Tissue: Blood / Cell: Plasma Cell / Location in cell: extracellular Homo sapiens (human) / synonym: Human / Tissue: Blood / Cell: Plasma Cell / Location in cell: extracellular |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50mM Tris-HCL |
| Grid | Details: C-flat 1.2/1.3-2C 400 mesh grids, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 50 % / Chamber temperature: 100 K / Instrument: GATAN CRYOPLUNGE 3 Method: Incubation of 0.014 mg/ml fibril solution on glow discharged holey carbon grid for 30 seconds and backside blotting for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 100 K |
| Date | Nov 12, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON I (4k x 4k) / Number real images: 10 / Average electron dose: 25 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 66350.7 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)