[English] 日本語
 Yorodumi
Yorodumi- EMDB-12181: Cryo-EM map of Icosahedrally averaged native core of Pyruvate Deh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12181 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Icosahedrally averaged native core of Pyruvate Dehydrogenase Complex from Ch. thermophilum | |||||||||
 Map data Map data | Symmetrically averaged (Icosahedral Symmetry) Pyruvate Dehydrogenase Complex core from Ch. thermophilum. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dihydrolipoyl / transacetylase / E2 / Pyruvate / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationdihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / pyruvate decarboxylation to acetyl-CoA / pyruvate dehydrogenase complex / mitochondrion Similarity search - Function | |||||||||
| Biological species |  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | |||||||||
 Authors Authors | Skalidis I / Kyrilis FL / Tueting C / Semchonok DA / Kastritis PL | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts. Authors: Fotis L Kyrilis / Dmitry A Semchonok / Ioannis Skalidis / Christian Tüting / Farzad Hamdi / Francis J O'Reilly / Juri Rappsilber / Panagiotis L Kastritis /   Abstract: The pyruvate dehydrogenase complex (PDHc) is a giant enzymatic assembly involved in pyruvate oxidation. PDHc components have been characterized in isolation, but the complex's quaternary structure ...The pyruvate dehydrogenase complex (PDHc) is a giant enzymatic assembly involved in pyruvate oxidation. PDHc components have been characterized in isolation, but the complex's quaternary structure has remained elusive due to sheer size, heterogeneity, and plasticity. Here, we identify fully assembled Chaetomium thermophilum α-keto acid dehydrogenase complexes in native cell extracts and characterize their domain arrangements utilizing mass spectrometry, activity assays, crosslinking, electron microscopy (EM), and computational modeling. We report the cryo-EM structure of the PDHc core and observe unique features of the previously unknown native state. The asymmetric reconstruction of the 10-MDa PDHc resolves spatial proximity of its components, agrees with stoichiometric data (60 E2p:12 E3BP:∼20 E1p: ≤ 12 E3), and proposes a minimum reaction path among component enzymes. The PDHc shows the presence of a dynamic pyruvate oxidation compartment, organized by core and peripheral protein species. Our data provide a framework for further understanding PDHc and α-keto acid dehydrogenase complex structure and function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12181.map.gz emd_12181.map.gz | 45.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12181-v30.xml emd-12181-v30.xml emd-12181.xml emd-12181.xml | 30.8 KB 30.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12181_fsc.xml emd_12181_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12181.png emd_12181.png | 64.6 KB | ||
| Filedesc metadata |  emd-12181.cif.gz emd-12181.cif.gz | 6.3 KB | ||
| Others |  emd_12181_additional_1.map.gz emd_12181_additional_1.map.gz emd_12181_additional_2.map.gz emd_12181_additional_2.map.gz emd_12181_additional_3.map.gz emd_12181_additional_3.map.gz emd_12181_additional_4.map.gz emd_12181_additional_4.map.gz emd_12181_additional_5.map.gz emd_12181_additional_5.map.gz emd_12181_additional_6.map.gz emd_12181_additional_6.map.gz emd_12181_half_map_1.map.gz emd_12181_half_map_1.map.gz emd_12181_half_map_2.map.gz emd_12181_half_map_2.map.gz | 46.2 MB 46.2 MB 46.2 MB 46.2 MB 46.2 MB 46.1 MB 45.9 MB 45.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12181 http://ftp.pdbj.org/pub/emdb/structures/EMD-12181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12181 | HTTPS FTP |
-Related structure data
| Related structure data |  7bgjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10625 (Title: Raw cryo- and negative stain micrographs of a native extract from C. thermophilum used to reconstruct oxo acid dehydrogenase complexes EMPIAR-10625 (Title: Raw cryo- and negative stain micrographs of a native extract from C. thermophilum used to reconstruct oxo acid dehydrogenase complexesData size: 5.4 TB Data #1: Unaligned multiframe and summed frame mrc files of the datasets presented in the corresponding Cell Reports manuscript, by Kastritis et al. Please read the README file for more information ...Data #1: Unaligned multiframe and summed frame mrc files of the datasets presented in the corresponding Cell Reports manuscript, by Kastritis et al. Please read the README file for more information [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12181.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12181.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetrically averaged (Icosahedral Symmetry) Pyruvate Dehydrogenase Complex core from Ch. thermophilum. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.1765 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Asymmetrical Pyruvate Dehydrogenase Complex from Ch. thermophilum.
| File | emd_12181_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetrical Pyruvate Dehydrogenase Complex from Ch. thermophilum. | ||||||||||||
| Projections & Slices |
| ||||||||||||
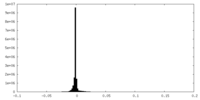
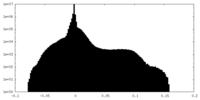
| Density Histograms |
-Additional map: Asymmetrical Pyruvate Dehydrogenase Complex from Ch. thermophilum, Half-map...
| File | emd_12181_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetrical Pyruvate Dehydrogenase Complex from Ch. thermophilum, Half-map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
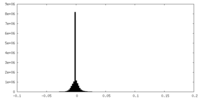
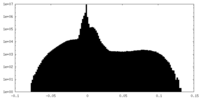
| Density Histograms |
-Additional map: Asymmetrical Pyruvate Dehydrogenase Complex from Ch. thermophilum, Half-map...
| File | emd_12181_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetrical Pyruvate Dehydrogenase Complex from Ch. thermophilum, Half-map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Asymmetrical Pyruvate Dehydrogenase Complex core from Ch. thermophilum....
| File | emd_12181_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetrical Pyruvate Dehydrogenase Complex core from Ch. thermophilum. Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Asymmetrical Pyruvate Dehydrogenase Complex core from Ch. thermophilum....
| File | emd_12181_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetrical Pyruvate Dehydrogenase Complex core from Ch. thermophilum. Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Asymmetrical Pyruvate Dehydrogenase Complex core from Ch. thermophilum....
| File | emd_12181_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetrical Pyruvate Dehydrogenase Complex core from Ch. thermophilum. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Symmetrically averaged (Icosahedral Symmetry) Pyruvate Dehydrogenase Complex core...
| File | emd_12181_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetrically averaged (Icosahedral Symmetry) Pyruvate Dehydrogenase Complex core from Ch. thermophilum, Half-map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Symmetrically averaged (Icosahedral Symmetry) Pyruvate Dehydrogenase Complex core...
| File | emd_12181_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetrically averaged (Icosahedral Symmetry) Pyruvate Dehydrogenase Complex core from Ch. thermophilum, Half-map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Native 60-mer core of Pyruvate Dehydrogenase Complex
| Entire | Name: Native 60-mer core of Pyruvate Dehydrogenase Complex |
|---|---|
| Components |
|
-Supramolecule #1: Native 60-mer core of Pyruvate Dehydrogenase Complex
| Supramolecule | Name: Native 60-mer core of Pyruvate Dehydrogenase Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 3 MDa |
-Macromolecule #1: Acetyltransferase component of pyruvate dehydrogenase complex
| Macromolecule | Name: Acetyltransferase component of pyruvate dehydrogenase complex type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: dihydrolipoyllysine-residue acetyltransferase |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 48.777488 KDa |
| Sequence | String: MLAQVLRRQA LQHVRLARAA APSLTRWYAS YPPHTIVKMP ALSPTMTSGN IGAWQKKPGD AITPGEVLVE IETDKAQMDF EFQEEGVLA KILKETGEKD VAVGSPIAVL VEEGTDINAF QNFTLEDAGG DAAAPAAPAK EELAKAETAP TPASTSAPEP E ETTSTGKL ...String: MLAQVLRRQA LQHVRLARAA APSLTRWYAS YPPHTIVKMP ALSPTMTSGN IGAWQKKPGD AITPGEVLVE IETDKAQMDF EFQEEGVLA KILKETGEKD VAVGSPIAVL VEEGTDINAF QNFTLEDAGG DAAAPAAPAK EELAKAETAP TPASTSAPEP E ETTSTGKL EPALDREPNV SFAAKKLAHE LDVPLKALKG TGPGGKITEE DVKKAASAPA AAAAAPGAAY QDIPISNMRK TI ATRLKES VSENPHFFVT SELSVSKLLK LRQALNSSAE GRYKLSVNDF LIKAIAVACK RVPAVNSSWR DGVIRQFDTV DVS VAVATP TGLITPIVKG VEAKGLETIS ATVKELAKKA RDGKLKPEDY QGGTISISNM GMNPAVERFT AIINPPQAAI LAVG TTKKV AVPVENEDGT TGVEWDDQIV VTASFDHKVV DGAVGAEWMR ELKKVVENPL ELLL UniProtKB: Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 200.0 mM / Component - Formula: NH4CH2COOH / Component - Name: Ammonium acetate |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: For plunging, blot force 2 and blotting time of 6 sec were applied.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 77.15 K / Max: 103.15 K |
| Alignment procedure | Coma free - Residual tilt: 14.7 mrad |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 2593 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 44067 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 444-647 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The initial model was used to generate our model using MODELLER. This inital model was fitted into the density using ChimeraX and refined in real space using PHENIX. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 103.25 |
| Output model |  PDB-7bgj: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)