+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11997 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a nanoparticle for a COVID-19 vaccine candidate | |||||||||
 Map data Map data | RBD Spy mi3 cage vlp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / SpyTag / VLP / Receptor-binding domain / vaccine / COVID-19 / SpyCatcher / nanocage / icosahedral / nanoparticle / coronavirus / mi3 / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationD-galactonate catabolic process / 2-dehydro-3-deoxy-6-phosphogalactonate aldolase activity / cell adhesion / extracellular region Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria) Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Duyvesteyn HME / Stuart DI | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Authors: Tiong Kit Tan / Pramila Rijal / Rolle Rahikainen / Anthony H Keeble / Lisa Schimanski / Saira Hussain / Ruth Harvey / Jack W P Hayes / Jane C Edwards / Rebecca K McLean / Veronica Martini / ...Authors: Tiong Kit Tan / Pramila Rijal / Rolle Rahikainen / Anthony H Keeble / Lisa Schimanski / Saira Hussain / Ruth Harvey / Jack W P Hayes / Jane C Edwards / Rebecca K McLean / Veronica Martini / Miriam Pedrera / Nazia Thakur / Carina Conceicao / Isabelle Dietrich / Holly Shelton / Anna Ludi / Ginette Wilsden / Clare Browning / Adrian K Zagrajek / Dagmara Bialy / Sushant Bhat / Phoebe Stevenson-Leggett / Philippa Hollinghurst / Matthew Tully / Katy Moffat / Chris Chiu / Ryan Waters / Ashley Gray / Mehreen Azhar / Valerie Mioulet / Joseph Newman / Amin S Asfor / Alison Burman / Sylvia Crossley / John A Hammond / Elma Tchilian / Bryan Charleston / Dalan Bailey / Tobias J Tuthill / Simon P Graham / Helen M E Duyvesteyn / Tomas Malinauskas / Jiandong Huo / Julia A Tree / Karen R Buttigieg / Raymond J Owens / Miles W Carroll / Rodney S Daniels / John W McCauley / David I Stuart / Kuan-Ying A Huang / Mark Howarth / Alain R Townsend /   Abstract: There is need for effective and affordable vaccines against SARS-CoV-2 to tackle the ongoing pandemic. In this study, we describe a protein nanoparticle vaccine against SARS-CoV-2. The vaccine is ...There is need for effective and affordable vaccines against SARS-CoV-2 to tackle the ongoing pandemic. In this study, we describe a protein nanoparticle vaccine against SARS-CoV-2. The vaccine is based on the display of coronavirus spike glycoprotein receptor-binding domain (RBD) on a synthetic virus-like particle (VLP) platform, SpyCatcher003-mi3, using SpyTag/SpyCatcher technology. Low doses of RBD-SpyVLP in a prime-boost regimen induce a strong neutralising antibody response in mice and pigs that is superior to convalescent human sera. We evaluate antibody quality using ACE2 blocking and neutralisation of cell infection by pseudovirus or wild-type SARS-CoV-2. Using competition assays with a monoclonal antibody panel, we show that RBD-SpyVLP induces a polyclonal antibody response that recognises key epitopes on the RBD, reducing the likelihood of selecting neutralisation-escape mutants. Moreover, RBD-SpyVLP is thermostable and can be lyophilised without losing immunogenicity, to facilitate global distribution and reduce cold-chain dependence. The data suggests that RBD-SpyVLP provides strong potential to address clinical and logistic challenges of the COVID-19 pandemic. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11997.map.gz emd_11997.map.gz | 66.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11997-v30.xml emd-11997-v30.xml emd-11997.xml emd-11997.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11997.png emd_11997.png | 178.9 KB | ||
| Filedesc metadata |  emd-11997.cif.gz emd-11997.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11997 http://ftp.pdbj.org/pub/emdb/structures/EMD-11997 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11997 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11997 | HTTPS FTP |
-Related structure data
| Related structure data |  7b3yMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11997.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11997.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RBD Spy mi3 cage vlp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.63 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
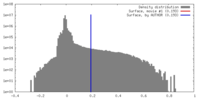
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RBD-Spycatcher-mi3
| Entire | Name: RBD-Spycatcher-mi3 |
|---|---|
| Components |
|
-Supramolecule #1: RBD-Spycatcher-mi3
| Supramolecule | Name: RBD-Spycatcher-mi3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate ald...
| Macromolecule | Name: Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase type: protein_or_peptide / ID: 1 Details: Model just contains mi3 cage component of VLP. Note that the Spy and RBD (spy tag and RBD are omitted from the sequence) are are not included in the model. The spycatcher003-mi3 can be found ...Details: Model just contains mi3 cage component of VLP. Note that the Spy and RBD (spy tag and RBD are omitted from the sequence) are are not included in the model. The spycatcher003-mi3 can be found on genbank with code MT945417 and that of the SpyTag-RBD at MT945427.,Model just contains mi3 cage component of VLP. Note that the Spy and RBD (spy tag and RBD are omitted from the sequence) are are not included in the model. The spycatcher003-mi3 can be found on genbank with code MT945417 and that of the SpyTag-RBD at MT945427. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria) Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria)Strain: ATCC 43589 / MSB8 / DSM 3109 / JCM 10099 |
| Molecular weight | Theoretical: 36.000715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSVTTLSG LSGEQGPSGD MTTEEDSATH IKFSKRDEDG RELAGATMEL RDSSGKTIST WISDGHVKDF YLYPGKYTFV ETAAPDGYE VATPIEFTVN EDGQVTVDGE ATEGDAHTGG SGGSGGSGGS MKMEELFKKH KIVAVLRANS VEEAKKKALA V FLGGVHLI ...String: MGSSVTTLSG LSGEQGPSGD MTTEEDSATH IKFSKRDEDG RELAGATMEL RDSSGKTIST WISDGHVKDF YLYPGKYTFV ETAAPDGYE VATPIEFTVN EDGQVTVDGE ATEGDAHTGG SGGSGGSGGS MKMEELFKKH KIVAVLRANS VEEAKKKALA V FLGGVHLI EITFTVPDAD TVIKELSFLK EMGAIIGAGT VTSVEQARKA VESGAEFIVS PHLDEEISQF AKEKGVFYMP GV MTPTELV KAMKLGHTIL KLFPGEVVGP QFVKAMKGPF PNVKFVPTGG VNLDNVCEWF KAGVLAVGVG SALVKGTPVE VAE KAKAFV EKIRGCTEGS GEPEA UniProtKB: Fibronectin binding protein, 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: PBS |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 7393 / Average electron dose: 48.1 e/Å2 Details: SerialEM collection employing beam tilt for multishots per hole using custom script. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 165000 / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)