[English] 日本語
 Yorodumi
Yorodumi- EMDB-8278: Structure of Nanoparticle Released from Enveloped Protein Nanoparticle -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8278 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Nanoparticle Released from Enveloped Protein Nanoparticle | ||||||||||||||||||
 Map data Map data | Icosahedrally-symmetrized, average reconstruction of nanoparticle released from EPN | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | protein design / icosahedral assemblies / cell transduction / enveloped viruses / virus assembly / enveloped protein / nanoparticle / STRUCTURAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationD-galactonate catabolic process / 2-dehydro-3-deoxy-6-phosphogalactonate aldolase activity / viral budding via host ESCRT complex / host multivesicular body / viral nucleocapsid / viral translational frameshifting / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity ...D-galactonate catabolic process / 2-dehydro-3-deoxy-6-phosphogalactonate aldolase activity / viral budding via host ESCRT complex / host multivesicular body / viral nucleocapsid / viral translational frameshifting / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / RNA binding / zinc ion binding / identical protein binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |   Thermotoga maritima (bacteria) / Thermotoga maritima (bacteria) /  Human immunodeficiency virus type 1 group M subtype B (isolate BH10) Human immunodeficiency virus type 1 group M subtype B (isolate BH10) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | ||||||||||||||||||
 Authors Authors | Votteler J / Ogohara C | ||||||||||||||||||
| Funding support |  Germany, Germany,  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Designed proteins induce the formation of nanocage-containing extracellular vesicles. Authors: Jörg Votteler / Cassandra Ogohara / Sue Yi / Yang Hsia / Una Nattermann / David M Belnap / Neil P King / Wesley I Sundquist /  Abstract: Complex biological processes are often performed by self-organizing nanostructures comprising multiple classes of macromolecules, such as ribosomes (proteins and RNA) or enveloped viruses (proteins, ...Complex biological processes are often performed by self-organizing nanostructures comprising multiple classes of macromolecules, such as ribosomes (proteins and RNA) or enveloped viruses (proteins, nucleic acids and lipids). Approaches have been developed for designing self-assembling structures consisting of either nucleic acids or proteins, but strategies for engineering hybrid biological materials are only beginning to emerge. Here we describe the design of self-assembling protein nanocages that direct their own release from human cells inside small vesicles in a manner that resembles some viruses. We refer to these hybrid biomaterials as 'enveloped protein nanocages' (EPNs). Robust EPN biogenesis requires protein sequence elements that encode three distinct functions: membrane binding, self-assembly, and recruitment of the endosomal sorting complexes required for transport (ESCRT) machinery. A variety of synthetic proteins with these functional elements induce EPN biogenesis, highlighting the modularity and generality of the design strategy. Biochemical analyses and cryo-electron microscopy reveal that one design, EPN-01, comprises small (~100 nm) vesicles containing multiple protein nanocages that closely match the structure of the designed 60-subunit self-assembling scaffold. EPNs that incorporate the vesicular stomatitis viral glycoprotein can fuse with target cells and deliver their contents, thereby transferring cargoes from one cell to another. These results show how proteins can be programmed to direct the formation of hybrid biological materials that perform complex tasks, and establish EPNs as a class of designed, modular, genetically-encoded nanomaterials that can transfer molecules between cells. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8278.map.gz emd_8278.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8278-v30.xml emd-8278-v30.xml emd-8278.xml emd-8278.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8278.png emd_8278.png | 196.4 KB | ||
| Filedesc metadata |  emd-8278.cif.gz emd-8278.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8278 http://ftp.pdbj.org/pub/emdb/structures/EMD-8278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8278 | HTTPS FTP |
-Validation report
| Summary document |  emd_8278_validation.pdf.gz emd_8278_validation.pdf.gz | 498.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8278_full_validation.pdf.gz emd_8278_full_validation.pdf.gz | 497.8 KB | Display | |
| Data in XML |  emd_8278_validation.xml.gz emd_8278_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_8278_validation.cif.gz emd_8278_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8278 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8278 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8278 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8278 | HTTPS FTP |
-Related structure data
| Related structure data |  5kp9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8278.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8278.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedrally-symmetrized, average reconstruction of nanoparticle released from EPN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.193 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
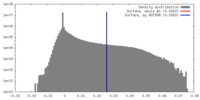
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EPN-01*
| Entire | Name: EPN-01* |
|---|---|
| Components |
|
-Supramolecule #1: EPN-01*
| Supramolecule | Name: EPN-01* / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
-Macromolecule #1: EPN-01*
| Macromolecule | Name: EPN-01* / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human immunodeficiency virus type 1 group M subtype B (isolate BH10) Human immunodeficiency virus type 1 group M subtype B (isolate BH10)Strain: isolate BH10 |
| Molecular weight | Theoretical: 30.30492 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGARASGSKS GSGSDSGSKM EELFKKHKIV AVLRANSVEE AKKKALAVFL GGVHLIEITF TVPDADTVIK ELSFLKEMGA IIGAGTVTS VEQCRKAVES GAEFIVSPHL DEEISQFCKE KGVFYMPGVM TPTELVKAMK LGHTILKLFP GEVVGPQFVK A MKGPFPNV ...String: MGARASGSKS GSGSDSGSKM EELFKKHKIV AVLRANSVEE AKKKALAVFL GGVHLIEITF TVPDADTVIK ELSFLKEMGA IIGAGTVTS VEQCRKAVES GAEFIVSPHL DEEISQFCKE KGVFYMPGVM TPTELVKAMK LGHTILKLFP GEVVGPQFVK A MKGPFPNV KFVPTGGVNL DNVCEWFKAG VLAVGVGSAL VKGTPVEVAE KAKAFVEKIR GCTEQKLISE EDLQSRPEPT AP PEESFRS GVETTTPPQK QEPIDKELYP LTSLRSLFGN DPSSQ UniProtKB: 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase, Gag polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: Phosphate-buffered saline |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II / Details: 11 second blot, 0 mm offset. |
| Details | EPN nanoparticles released from vesicles by detergent treatment (0.75% CHAPS) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Average electron dose: 2.0 e/Å2 / Details: Electron dose at specimen was not recorded. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.3000000000000003 µm / Calibrated defocus min: 0.7000000000000001 µm / Calibrated magnification: 41911 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5kp9: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)