[English] 日本語
 Yorodumi
Yorodumi- EMDB-11799: Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11799 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Reference Map) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CCC / transporter / human membrane protein / homodimer / KCC3 / KCC / potassium-chloride coupled transporter / Structural Genomics / Structural Genomics Consortium / SGC / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC12A6 causes agenesis of the corpus callosum, with peripheral neuropathy (ACCPN) / potassium ion transmembrane transporter activity / potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / chloride ion homeostasis / cellular hypotonic response / cellular hypotonic salinity response / potassium ion homeostasis / cell volume homeostasis / potassium ion import across plasma membrane ...Defective SLC12A6 causes agenesis of the corpus callosum, with peripheral neuropathy (ACCPN) / potassium ion transmembrane transporter activity / potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / chloride ion homeostasis / cellular hypotonic response / cellular hypotonic salinity response / potassium ion homeostasis / cell volume homeostasis / potassium ion import across plasma membrane / monoatomic ion transport / chloride transmembrane transport / potassium ion transmembrane transport / cellular response to glucose stimulus / basolateral plasma membrane / chemical synaptic transmission / angiogenesis / axon / synapse / protein kinase binding / membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
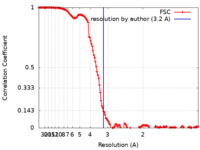
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Chi G / Man H | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2021 Journal: EMBO J / Year: 2021Title: Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters. Authors: Gamma Chi / Rebecca Ebenhoch / Henry Man / Haiping Tang / Laurence E Tremblay / Gabriella Reggiano / Xingyu Qiu / Tina Bohstedt / Idlir Liko / Fernando G Almeida / Alexandre P Garneau / Dong ...Authors: Gamma Chi / Rebecca Ebenhoch / Henry Man / Haiping Tang / Laurence E Tremblay / Gabriella Reggiano / Xingyu Qiu / Tina Bohstedt / Idlir Liko / Fernando G Almeida / Alexandre P Garneau / Dong Wang / Gavin McKinley / Christophe P Moreau / Kiran D Bountra / Patrizia Abrusci / Shubhashish M M Mukhopadhyay / Alejandra Fernandez-Cid / Samira Slimani / Julie L Lavoie / Nicola A Burgess-Brown / Ben Tehan / Frank DiMaio / Ali Jazayeri / Paul Isenring / Carol V Robinson / Katharina L Dürr /    Abstract: Potassium-coupled chloride transporters (KCCs) play crucial roles in regulating cell volume and intracellular chloride concentration. They are characteristically inhibited under isotonic conditions ...Potassium-coupled chloride transporters (KCCs) play crucial roles in regulating cell volume and intracellular chloride concentration. They are characteristically inhibited under isotonic conditions via phospho-regulatory sites located within the cytoplasmic termini. Decreased inhibitory phosphorylation in response to hypotonic cell swelling stimulates transport activity, and dysfunction of this regulatory process has been associated with various human diseases. Here, we present cryo-EM structures of human KCC3b and KCC1, revealing structural determinants for phospho-regulation in both N- and C-termini. We show that phospho-mimetic KCC3b is arrested in an inward-facing state in which intracellular ion access is blocked by extensive contacts with the N-terminus. In another mutant with increased isotonic transport activity, KCC1Δ19, this interdomain interaction is absent, likely due to a unique phospho-regulatory site in the KCC1 N-terminus. Furthermore, we map additional phosphorylation sites as well as a previously unknown ATP/ADP-binding pocket in the large C-terminal domain and show enhanced thermal stabilization of other CCCs by adenine nucleotides. These findings provide fundamentally new insights into the complex regulation of KCCs and may unlock innovative strategies for drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11799.map.gz emd_11799.map.gz | 229.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11799-v30.xml emd-11799-v30.xml emd-11799.xml emd-11799.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11799_fsc.xml emd_11799_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11799.png emd_11799.png | 161.3 KB | ||
| Masks |  emd_11799_msk_1.map emd_11799_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11799.cif.gz emd-11799.cif.gz | 6.9 KB | ||
| Others |  emd_11799_half_map_1.map.gz emd_11799_half_map_1.map.gz emd_11799_half_map_2.map.gz emd_11799_half_map_2.map.gz | 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11799 http://ftp.pdbj.org/pub/emdb/structures/EMD-11799 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11799 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11799 | HTTPS FTP |
-Validation report
| Summary document |  emd_11799_validation.pdf.gz emd_11799_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11799_full_validation.pdf.gz emd_11799_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_11799_validation.xml.gz emd_11799_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_11799_validation.cif.gz emd_11799_validation.cif.gz | 28.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11799 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11799 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11799 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11799 | HTTPS FTP |
-Related structure data
| Related structure data |  7ainMC  6y5vC  7aioC  7aipC  7aiqC  7airC  7ngbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11799.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11799.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.651 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
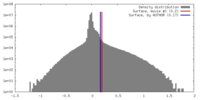
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11799_msk_1.map emd_11799_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11799_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11799_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimeric complex of human potassium chloride transporter KCC3b
| Entire | Name: Homodimeric complex of human potassium chloride transporter KCC3b |
|---|---|
| Components |
|
-Supramolecule #1: Homodimeric complex of human potassium chloride transporter KCC3b
| Supramolecule | Name: Homodimeric complex of human potassium chloride transporter KCC3b type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: Isoform 2 of Solute carrier family 12 member 6
| Macromolecule | Name: Isoform 2 of Solute carrier family 12 member 6 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 122.433297 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPHFTVTKVE DPEEGAAASI SQEPSLADIK ARIQDSDEPD LSQNDITGEH SQLLDDGHKK ARNAYLNNSN YEEGDEYFDK NLALFEEEM DTRPKVSSLL NRMANYTNLT QGAKEHEEAE NITEGKKKPT KTPQMGTFMG VYLPCLQNIF GVILFLRLTW V VGTAGVLQ ...String: MPHFTVTKVE DPEEGAAASI SQEPSLADIK ARIQDSDEPD LSQNDITGEH SQLLDDGHKK ARNAYLNNSN YEEGDEYFDK NLALFEEEM DTRPKVSSLL NRMANYTNLT QGAKEHEEAE NITEGKKKPT KTPQMGTFMG VYLPCLQNIF GVILFLRLTW V VGTAGVLQ AFAIVLICCC CTMLTAISMS AIATNGVVPA GGSYFMISRA LGPEFGGAVG LCFYLGTTFA AAMYILGAIE IF LVYIVPR AAIFHSDDAL KESAAMLNNM RVYGTAFLVL MVLVVFIGVR YVNKFASLFL ACVIVSILAI YAGAIKSSFA PPH FPVCML GNRTLSSRHI DVCSKTKEIN NMTVPSKLWG FFCNSSQFFN ATCDEYFVHN NVTSIQGIPG LASGIITENL WSNY LPKGE IIEKPSAKSS DVLGSLNHEY VLVDITTSFT LLVGIFFPSV TGIMAGSNRS GDLKDAQKSI PIGTILAILT TSFVY LSNV VLFGACIEGV VLRDKFGDAV KGNLVVGTLS WPSPWVIVIG SFFSTCGAGL QSLTGAPRLL QAIAKDNIIP FLRVFG HSK ANGEPTWALL LTAAIAELGI LIASLDLVAP ILSMFFLMCY LFVNLACALQ TLLRTPNWRP RFRYYHWALS FMGMSIC LA LMFISSWYYA IVAMVIAGMI YKYIEYQGAE KEWGDGIRGL SLSAARFALL RLEEGPPHTK NWRPQLLVLL KLDEDLHV K HPRLL(TPO)FASQ LKAGKGLTIV GSVIVGNFLE NYGEALAAEQ TIKHLMEAEK VKGFCQLVVA AKLREGISHL IQSCG LGGM KHNTVVMGWP NGWRQSEDAR AWKTFIGTVR VTTAAHLALL VAKNISFFPS NVEQFSEGNI DVWWIVHDGG MLMLLP FLL KQHKVWRKCS IRIFTVAQLE DNSIQMKKDL ATFLYHLRIE AEVEVVEMHD (SEP)DISAYTYER DLMMEQRSQM LRH MRLSKT ERDREAQLVK DRNSMLRLTS IGSDEDEETE TYQEKVHMDW TKDKYMASRG QKAKSMEGFQ DLLNMRPDQS NVRR MHTAV KLNEVIVNKS HEAKLVLLNM PGPPRNPEGD ENYMEFLEVL TEGLERVLLV RGGGSEVITI YS UniProtKB: Solute carrier family 12 member 6 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Model from previous map/model repositioned with Chimera and refined with Coot/Phenix. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-7ain: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)