+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10703 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Human Potassium Chloride Transporter KCC3 in NaCl | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dimer / transporter / potassium chloride / KCC3 / SLC12A6 / membrane protein / APC / Structural Genomics / Structural Genomics Consortium / SGC / PSI-2 / Protein Structure Initiative | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC12A6 causes agenesis of the corpus callosum, with peripheral neuropathy (ACCPN) / potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / potassium ion transmembrane transporter activity / chloride ion homeostasis / cellular hypotonic salinity response / cellular hypotonic response / potassium ion homeostasis / cell volume homeostasis / potassium ion import across plasma membrane ...Defective SLC12A6 causes agenesis of the corpus callosum, with peripheral neuropathy (ACCPN) / potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / potassium ion transmembrane transporter activity / chloride ion homeostasis / cellular hypotonic salinity response / cellular hypotonic response / potassium ion homeostasis / cell volume homeostasis / potassium ion import across plasma membrane / monoatomic ion transport / potassium ion transmembrane transport / chloride transmembrane transport / cellular response to glucose stimulus / angiogenesis / basolateral plasma membrane / chemical synaptic transmission / axon / synapse / protein kinase binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
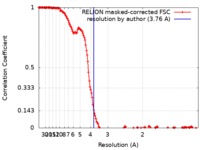
| Method | single particle reconstruction / cryo EM / Resolution: 3.76 Å | |||||||||
 Authors Authors | Chi G / Man H | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Structure of Human Potassium Chloride Transporter KCC3 in NaCl Authors: Chi G / Man H / Ebenhoch R / Reggiano G / Pike ACW / Wang D / McKinley G / Mukhopadhyay SMM / Chalk R / Moreau C / Snee M / Bohstedt T / Singh NK / Abrusci P / Arrowsmith CH / Bountra C / ...Authors: Chi G / Man H / Ebenhoch R / Reggiano G / Pike ACW / Wang D / McKinley G / Mukhopadhyay SMM / Chalk R / Moreau C / Snee M / Bohstedt T / Singh NK / Abrusci P / Arrowsmith CH / Bountra C / Edwards AM / Marsden BD / Burgess-Brown NA / DiMaio F / Duerr KL / Structural Genomics Consortium (SGC) | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10703.map.gz emd_10703.map.gz | 304.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10703-v30.xml emd-10703-v30.xml emd-10703.xml emd-10703.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10703_fsc.xml emd_10703_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10703.png emd_10703.png | 156.6 KB | ||
| Filedesc metadata |  emd-10703.cif.gz emd-10703.cif.gz | 6.7 KB | ||
| Others |  emd_10703_half_map_1.map.gz emd_10703_half_map_1.map.gz emd_10703_half_map_2.map.gz emd_10703_half_map_2.map.gz | 304.1 MB 304.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10703 http://ftp.pdbj.org/pub/emdb/structures/EMD-10703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10703 | HTTPS FTP |
-Related structure data
| Related structure data |  6y5rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10703.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10703.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.651 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_10703_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
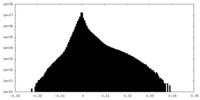
| Projections & Slices |
| ||||||||||||
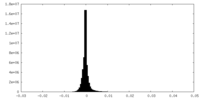
| Density Histograms |
-Half map: #2
| File | emd_10703_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
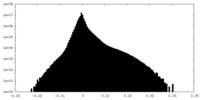
| Projections & Slices |
| ||||||||||||
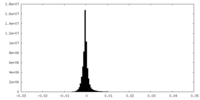
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimeric complex of human potassium chloride transporter
| Entire | Name: Homodimeric complex of human potassium chloride transporter |
|---|---|
| Components |
|
-Supramolecule #1: Homodimeric complex of human potassium chloride transporter
| Supramolecule | Name: Homodimeric complex of human potassium chloride transporter type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 246 KDa |
-Macromolecule #1: Solute carrier family 12 member 6
| Macromolecule | Name: Solute carrier family 12 member 6 / type: protein_or_peptide / ID: 1 Details: Sugar molecules from glycosylation post-translational modifications (NAG, BMA) present. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 122.273344 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPHFTVTKVE DPEEGAAASI SQEPSLADIK ARIQDSDEPD LSQNDITGEH SQLLDDGHKK ARNAYLNNSN YEEGDEYFDK NLALFEEEM DTRPKVSSLL NRMANYTNLT QGAKEHEEAE NITEGKKKPT KTPQMGTFMG VYLPCLQNIF GVILFLRLTW V VGTAGVLQ ...String: MPHFTVTKVE DPEEGAAASI SQEPSLADIK ARIQDSDEPD LSQNDITGEH SQLLDDGHKK ARNAYLNNSN YEEGDEYFDK NLALFEEEM DTRPKVSSLL NRMANYTNLT QGAKEHEEAE NITEGKKKPT KTPQMGTFMG VYLPCLQNIF GVILFLRLTW V VGTAGVLQ AFAIVLICCC CTMLTAISMS AIATNGVVPA GGSYFMISRA LGPEFGGAVG LCFYLGTTFA AAMYILGAIE IF LVYIVPR AAIFHSDDAL KESAAMLNNM RVYGTAFLVL MVLVVFIGVR YVNKFASLFL ACVIVSILAI YAGAIKSSFA PPH FPVCML GNRTLSSRHI DVCSKTKEIN NMTVPSKLWG FFCNSSQFFN ATCDEYFVHN NVTSIQGIPG LASGIITENL WSNY LPKGE IIEKPSAKSS DVLGSLNHEY VLVDITTSFT LLVGIFFPSV TGIMAGSNRS GDLKDAQKSI PIGTILAILT TSFVY LSNV VLFGACIEGV VLRDKFGDAV KGNLVVGTLS WPSPWVIVIG SFFSTCGAGL QSLTGAPRLL QAIAKDNIIP FLRVFG HSK ANGEPTWALL LTAAIAELGI LIASLDLVAP ILSMFFLMCY LFVNLACALQ TLLRTPNWRP RFRYYHWALS FMGMSIC LA LMFISSWYYA IVAMVIAGMI YKYIEYQGAE KEWGDGIRGL SLSAARFALL RLEEGPPHTK NWRPQLLVLL KLDEDLHV K HPRLLTFASQ LKAGKGLTIV GSVIVGNFLE NYGEALAAEQ TIKHLMEAEK VKGFCQLVVA AKLREGISHL IQSCGLGGM KHNTVVMGWP NGWRQSEDAR AWKTFIGTVR VTTAAHLALL VAKNISFFPS NVEQFSEGNI DVWWIVHDGG MLMLLPFLLK QHKVWRKCS IRIFTVAQLE DNSIQMKKDL ATFLYHLRIE AEVEVVEMHD SDISAYTYER DLMMEQRSQM LRHMRLSKTE R DREAQLVK DRNSMLRLTS IGSDEDEETE TYQEKVHMDW TKDKYMASRG QKAKSMEGFQ DLLNMRPDQS NVRRMHTAVK LN EVIVNKS HEAKLVLLNM PGPPRNPEGD ENYMEFLEVL TEGLERVLLV RGGGSEVITI YS UniProtKB: Solute carrier family 12 member 6 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV Details: 2 second blotting time, 40 second waiting time, -15 blotting force. |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 16472 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.3000000000000003 µm / Nominal defocus min: -0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)