[English] 日本語
 Yorodumi
Yorodumi- EMDB-11210: Structure of DPS determined by movement-free cryoEM with zero dos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11210 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of DPS determined by movement-free cryoEM with zero dose extrapolation | |||||||||
 Map data Map data | Masked map obtained suing zero-dose extrapolation thru exponential firs to amplitudes and linear fit to phases. The map is sharpened by a B factor of -55 Angstrom^2. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA-BINDING PROTEIN / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDnaA-Dps complex / Oxidoreductases; Oxidizing metal ions / oxidoreductase activity, acting on metal ions / nucleoid / chromosome condensation / response to starvation / response to stress / negative regulation of DNA-templated DNA replication initiation / ferric iron binding / intracellular iron ion homeostasis ...DnaA-Dps complex / Oxidoreductases; Oxidizing metal ions / oxidoreductase activity, acting on metal ions / nucleoid / chromosome condensation / response to starvation / response to stress / negative regulation of DNA-templated DNA replication initiation / ferric iron binding / intracellular iron ion homeostasis / DNA binding / identical protein binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | |||||||||
 Authors Authors | Naydenova K / Russo CJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Cryo-EM with sub-1 Å specimen movement. Authors: Katerina Naydenova / Peipei Jia / Christopher J Russo /   Abstract: Most information loss in cryogenic electron microscopy (cryo-EM) stems from particle movement during imaging, which remains poorly understood. We show that this movement is caused by buckling and ...Most information loss in cryogenic electron microscopy (cryo-EM) stems from particle movement during imaging, which remains poorly understood. We show that this movement is caused by buckling and subsequent deformation of the suspended ice, with a threshold that depends directly on the shape of the frozen water layer set by the support foil. We describe a specimen support design that eliminates buckling and reduces electron beam-induced particle movement to less than 1 angstrom. The design allows precise foil tracking during imaging with high-speed detectors, thereby lessening demands on cryostage precision and stability. It includes a maximal density of holes, which increases throughput in automated cryo-EM without degrading data quality. Movement-free imaging allows extrapolation to a three-dimensional map of the specimen at zero electron exposure, before the onset of radiation damage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Related structure data
| Related structure data |  6zglMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10445 (Title: Movies of DPS in 260 nm gold foil holes, which eliminate specimen movement EMPIAR-10445 (Title: Movies of DPS in 260 nm gold foil holes, which eliminate specimen movementData size: 1.5 TB Data #1: Unaligned multi-frame micrographs of DPS in 260 nm hole supports [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11210.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11210.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked map obtained suing zero-dose extrapolation thru exponential firs to amplitudes and linear fit to phases. The map is sharpened by a B factor of -55 Angstrom^2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
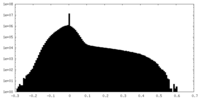
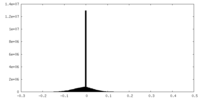
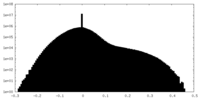
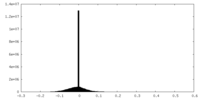
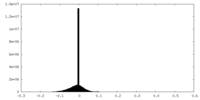
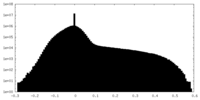
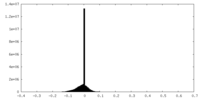
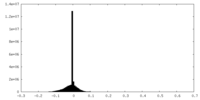
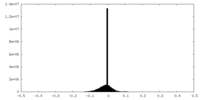
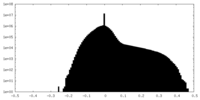
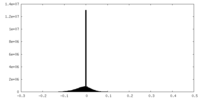
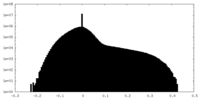
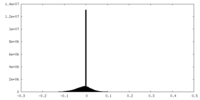
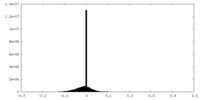
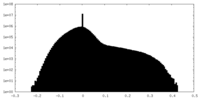
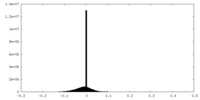
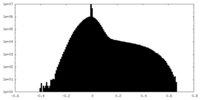
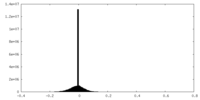
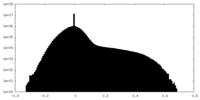
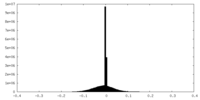
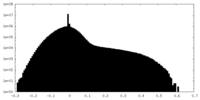
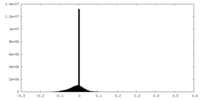
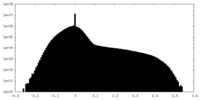
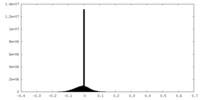
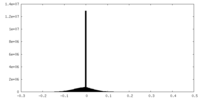
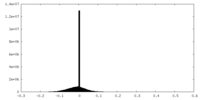
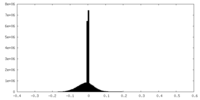
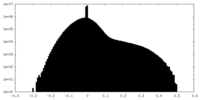
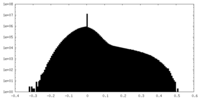
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.646 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
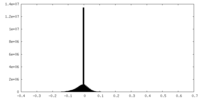
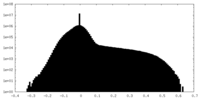
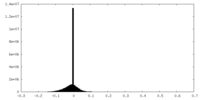
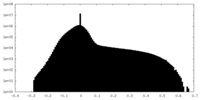
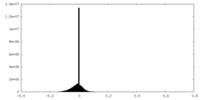
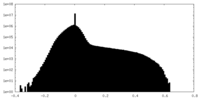
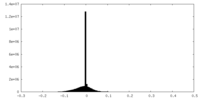
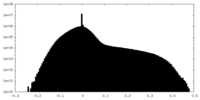
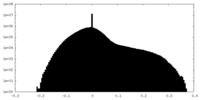
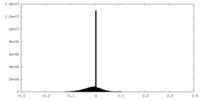
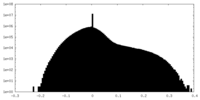
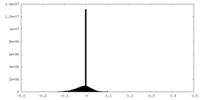
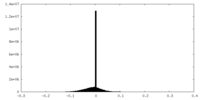
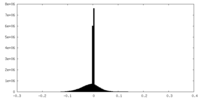
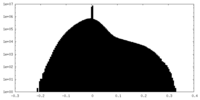
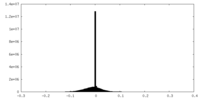
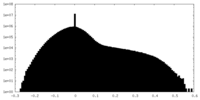
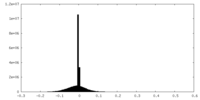
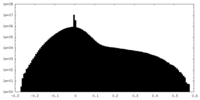
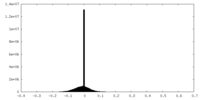
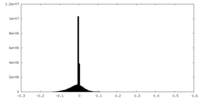
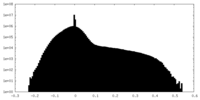
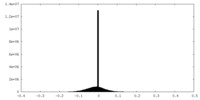
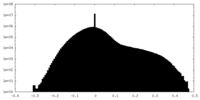
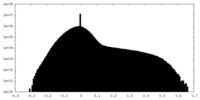
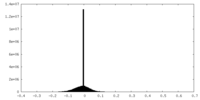
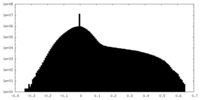
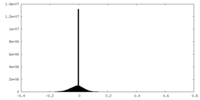
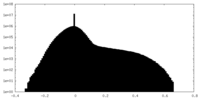
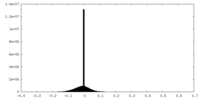
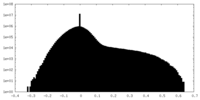
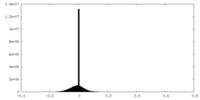
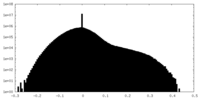
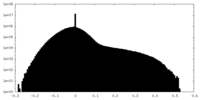
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Additional map: Frame 6 map
+Additional map: Frame 11 map
+Additional map: Frame 23 - half map 1
+Additional map: Frame 23 - half map 2
+Additional map: Frame 22 - half map 1
+Additional map: Frame 22 - half map 2
+Additional map: Frame 25 - half map 1
+Additional map: Frame 10 map
+Additional map: Frame 7 map
+Additional map: Frame 5 map
+Additional map: Frame 9 map
+Additional map: Frame 8 map
+Additional map: Frame 1 map
+Additional map: Frame 2 map
+Additional map: Frame 4 map
+Additional map: Frame 3 map
+Additional map: Frame 19 map
+Additional map: Frame 20 map
+Additional map: Frame 21 map
+Additional map: Frame 22 map
+Additional map: Frame 23 map
+Additional map: Frame 24 map
+Additional map: Frame 25 map
+Additional map: Frame 26 map
+Additional map: Frame 27 map
+Additional map: Frame 28 map
+Additional map: Frame 29 map
+Additional map: Frame 18 map
+Additional map: Frame 30 map
+Additional map: Frame 31 map
+Additional map: Frame 32 map
+Additional map: Frame 33 map
+Additional map: Frame 34 map
+Additional map: Same as main map, but sharpened with a...
+Additional map: Same as main map, but sharpened with a...
+Additional map: Frame 1 - half map 2
+Additional map: Frame 27 - half map 2
+Additional map: Frame 27 - half map 1
+Additional map: Frame 12 map
+Additional map: Frame 15 - half map 2
+Additional map: Frame 16 - half map 1
+Additional map: Frame 14 - half map 2
+Additional map: Frame 15 - half map 1
+Additional map: Frame 13 - half map 2
+Additional map: Frame 14 - half map 1
+Additional map: Frame 12 - half map 2
+Additional map: Frame 34 - half map 1
+Additional map: Frame 11 - half map 2
+Additional map: Frame 12 - half map 1
+Additional map: Frame 17 map
+Additional map: Frame 32 - half map 1
+Additional map: Frame 31 - half map 2
+Additional map: Frame 33 - half map 1
+Additional map: Frame 32 - half map 2
+Additional map: Frame 30 - half map 1
+Additional map: Frame 29 - half map 2
+Additional map: Frame 31 - half map 1
+Additional map: Frame 30 - half map 2
+Additional map: Frame 28 - half map 2
+Additional map: Frame 28 - half map 1
+Additional map: Frame 16 map
+Additional map: Frame 2 - half map 2
+Additional map: Frame 3 - half map 1
+Additional map: Frame 2 - half map 1
+Additional map: Frame 33 - half map 2
+Additional map: Frame 4 - half map 2
+Additional map: Frame 5 - half map 1
+Additional map: Frame 5 - half map 2
+Additional map: Frame 6 - half map 1
+Additional map: Frame 17 - half map 2
+Additional map: Frame 24 - half map 1
+Additional map: Frame 15 map
+Additional map: Frame 21 - half map 2
+Additional map: Frame 21 - half map 1
+Additional map: Frame 20 - half map 2
+Additional map: Frame 20 - half map 1
+Additional map: Frame 19 - half map 2
+Additional map: Frame 19 - half map 1
+Additional map: Frame 29 - half map 1
+Additional map: Frame 26 - half map 2
+Additional map: Frame 17 - half map 1
+Additional map: Frame 11 - half map 1
+Additional map: Frame 14 map
+Additional map: Frame 7 - half map 2
+Additional map: Frame 8 - half map 1
+Additional map: Frame 6 - half map 2
+Additional map: Frame 7 - half map 1
+Additional map: Frame 9 - half map 2
+Additional map: Frame 10 - half map 1
+Additional map: Frame 8 - half map 2
+Additional map: Frame 9 - half map 1
+Additional map: Frame 3 - half map 2
+Additional map: Frame 4 - half map 1
+Additional map: Frame 13 map
+Additional map: Frame 34 - half map 2
+Additional map: Frame 26 - half map 1
+Additional map: Frame 10 - half map 2
+Additional map: Frame 1 - half map 1
+Additional map: Frame 24 - half map 2
+Additional map: Frame 13 - half map 1
+Additional map: Frame 25 - half map 2
+Additional map: Frame 16 - half map 2
+Additional map: Frame 18 - half map 2
+Additional map: Frame 18 - half map 1
- Sample components
Sample components
-Entire : DNA protection during starvation protein (DPS)
| Entire | Name: DNA protection during starvation protein (DPS) |
|---|---|
| Components |
|
-Supramolecule #1: DNA protection during starvation protein (DPS)
| Supramolecule | Name: DNA protection during starvation protein (DPS) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA protection during starvation protein
| Macromolecule | Name: DNA protection during starvation protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.720295 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTAKLVKSK ATNLLYTRND VSDSEKKATV ELLNRQVIQF IDLSLITKQA HWNMRGANFI AVHEMLDGFR TALIDHLDTM AERAVQLGG VALGTTQVIN SKTPLKSYPL DIHNVQDHLK ELADRYAIVA NDVRKAIGEA KDDDTADILT AASRDLDKFL W FIESNIE UniProtKB: DNA protection during starvation protein |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 964 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Applied symmetry - Point group: T (tetrahedral) / Resolution.type: BY AUTHOR / Resolution: 1.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 275623 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)